Difference between revisions of "Documentation/Nightly/FAQ/Registration"
m (Text replacement - "https?:\/\/(?:www|wiki)\.slicer\.org\/slicerWiki\/index\.php\/([^ ]+) " to "https://www.slicer.org/wiki/$1 ") |
|||
| (28 intermediate revisions by 5 users not shown) | |||
| Line 2: | Line 2: | ||
<noinclude>__TOC__ | <noinclude>__TOC__ | ||
={{#titleparts: {{PAGENAME}} | | -1 }}=</noinclude><includeonly> | ={{#titleparts: {{PAGENAME}} | | -1 }}=</noinclude><includeonly> | ||
| − | ='''User FAQ: {{{1}}}'''= | + | {{#ifeq: {{#titleparts: {{PAGENAME}} | 3 }} | Documentation/{{documentation/version}}/Developers | | ='''User FAQ: {{{1}}}'''=}} |
</includeonly> | </includeonly> | ||
| Line 42: | Line 42: | ||
:5. alternatively if the entire orientation is wrong, i.e. coronal slices appear in the axial view etc., you may easier just change the ''space'' field to the proper orientation. Note that Slicer uses ''RAS'' space by default, i.e. first (x) axis = left-'''r'''ight, second (y) axis = posterior-'''a'''nterior, third (z) axis = inferior-'''s'''uperior | :5. alternatively if the entire orientation is wrong, i.e. coronal slices appear in the axial view etc., you may easier just change the ''space'' field to the proper orientation. Note that Slicer uses ''RAS'' space by default, i.e. first (x) axis = left-'''r'''ight, second (y) axis = posterior-'''a'''nterior, third (z) axis = inferior-'''s'''uperior | ||
:6. save & close the edited ''.nhdr'' file and reload the image in slicer to see if the orientation is now correct. | :6. save & close the edited ''.nhdr'' file and reload the image in slicer to see if the orientation is now correct. | ||
| + | === Can I undo the "centering" of an image === | ||
| + | When importing images, there's a "Center" checkbox, which if checked will reset the image origin to the center of the image grid, and ignore the image origin stored in the header. The same function is available to loaded images in the [[Documentation/{{documentation/version}}/Modules/Volumes|''Volumes'' module]] (Volume Information Tab). Results derived from images have their spatial info stored '''relative''' to that image origin. So fiducial points or label maps obtained from centered images will also be centered, which means they will align with a centered version of the image but not the original one. '''Is there a way to return such data to the original ''uncentered'' position?''' <br> | ||
| + | There is no dedicated module or function for that purpose currently implemented, but there are several ways to return data to the position before centering, provided the original image with the old origin is still available. Options are: | ||
| + | *copy the image origin (or entire spatial orientation info) from the original reference image into the header of the "centered" image. For images stored in a format where the header data is accessible in text format this is fairly straightforward. For other formats with binary headers it will require dedicated software to read the header and re-save the image. | ||
| + | *create a transform that embodies the shift and apply it to the data. This is probably the most accessible solution. It will work for all forms of data, i.e. fiducial points, labelmaps, surface models etc. To manually obtain such a transform: | ||
| + | #Load both original and centered image | ||
| + | #Go to the [[Documentation/{{documentation/version}}/Modules/Volumes|''Volumes'' module]], open the Volume Information Tab, then select either image and record the "Image Origin" information displayed. Calculate the difference of the two origins (x1-x2, y1-y2, z1-z2). | ||
| + | #Go to the [[Documentation/{{documentation/version}}/Modules/Transforms|''Transforms'' module]] , create a new transform, then enter the origin difference calculated above into the fields for translation (LR= left-right, PA=posterior-anterios, IS=inferior-superior). Note that to replicate the effect of centring the translation vector is centered-uncentered; to go back from centered to uncentered is the inverse of that transform, i.e. uncentered-centered. The "Invert Transform" button in the transforms module lets you switch between the two. | ||
| + | #Go to the [[Documentation/{{documentation/version}}/Modules/Data|''Data'' module]]. Drag the centered image and any data derived from it '''into'' the transform. | ||
| + | #Set fore- & background to original and centered image to verify, set the fade slider halfway so you can see both images. Verify that the two images align once the "centered" image has been placed into the transform. | ||
| + | #Right click on the meta data (e.g. fiducials) inside the transform, select '''Harden Transform''' from the pulldown menu. The data node will move back out of the transform to the main level to indicate the transform has been applied. | ||
| + | #Rename the data node (double click) to document that it has shifted. Then save it as a new file. | ||
| + | [[Media:Slicer_UncenterFiducials.mov|'''See here for a screencast describing this procedure to "uncenter" and shift meta data back to the original position''']]. <br> | ||
| + | For large sets of images calculating the offset manually may not be feasible. Below is a rudimentary python script that will read two or more images (NIfTI format) , calculate the offset and save it as an ITK transform (.tfm) file. You can then import this transform into Slicer and apply it to the data that needs shifting. | ||
| + | *As another alternative you can also run a quick automated registration of the centered to the uncentered image to obtain a transform. Note that this will look for a match based on image similarity and will not be 100% precise, but likely very close. | ||
| + | #! /usr/bin/env python | ||
| + | # reads 2 or more NIfTI images and extracts the image center offset to the first as an ITK transform file v1.0 | ||
| + | # usage: NIICenterOffset2ITK.py RefImg.nii CenteredImg1.nii CenteredImg2.nii ... | ||
| + | # output: CenteredImg1_center.tfm CenteredImg2_center.tfm ... | ||
| + | |||
| + | import nibabel as nib | ||
| + | import numpy | ||
| + | import sys | ||
| + | |||
| + | refimg = nib.load(sys.argv[1]) | ||
| + | refhdr = refimg.get_header() | ||
| + | reforigin=refhdr.get_qform()[([0,1,2],3)].astype(float) | ||
| + | for aImgName in sys.argv[2:]: | ||
| + | img = nib.load(aImgName) | ||
| + | hdr = img.get_header() | ||
| + | origin=hdr.get_qform()[([0,1,2],3)].astype(float) | ||
| + | offset=reforigin-origin | ||
| + | itk_file = open(aImgName+'.centered.tfm', "w") | ||
| + | itk_file.write('#Insight Transform File V1.0\n#Transform 0\nTransform: AffineTransform_double_3_3\n' ) | ||
| + | itk_file.write('Parameters: 1 0 0 0 1 0 0 0 1 %f %f %f\n' %( tuple( (reforigin-origin).tolist())) ) | ||
| + | itk_file.write('FixedParameters: 0 0 0\n') | ||
| + | itk_file.close() | ||
=== I have some DICOM images that I want to reslice at an arbitrary angle === | === I have some DICOM images that I want to reslice at an arbitrary angle === | ||
| Line 61: | Line 98: | ||
# Save the transform as a .tfm file. | # Save the transform as a .tfm file. | ||
# Inspect the contents of the .tfm file in a text editor, and compare them to what is shown in the 4x4 matrix in the Transforms module. | # Inspect the contents of the .tfm file in a text editor, and compare them to what is shown in the 4x4 matrix in the Transforms module. | ||
| − | # re-load the .tfm back into slicer and confirm you have the same data as you saved from | + | # re-load the .tfm back into slicer and confirm you have the same data as you saved from Slicer. |
| − | + | ||
| − | + | You will notice that the original and re-loaded Transforms are identical, but do not match the content of the .tfm file. The issue relates to the difference between Slicer, which uses a "computer graphics" view of the world, and ITK, which uses an "image processing" view of the world. The Slicer transform hierarchy models movement of an object from one spot to another. For example, a transform that has a positive "superior" value wrapped around a volume moves the volume up in patient space.<br> | |
| − | In | + | |
| + | Conversely, ITK transforms map "backwards": from the display space back to the original image. Imagine stepping sequentially through the output pixels: ITK wants to know the transform back to the input pixels ''used to calculate the output''. Additionally ITK transforms are saved in LPS, whereas the Slicer Transform widget uses RAS coordinates.<br> | ||
| + | |||
| + | In Summary: | ||
#The transform represented in the widget is in RAS. | #The transform represented in the widget is in RAS. | ||
#The transform represented in the tfm file is in LPS. | #The transform represented in the tfm file is in LPS. | ||
| − | #The transform represented in the file is the inverse of the transform in the widget | + | #The transform represented in the file is the inverse of the transform in the widget, and has the LPS/RAS conversion applied. |
#The order of the parameters in the tfm are the elements of the upper 3x3 of the transform displayed in the widget followed by the elements in the last column of the widget. | #The order of the parameters in the tfm are the elements of the upper 3x3 of the transform displayed in the widget followed by the elements in the last column of the widget. | ||
| − | + | ||
| − | + | Please see this discussion for more information, and code examples: | |
| − | + | ||
| − | + | https://www.slicer.org/wiki/Documentation/Nightly/Modules/Transforms#Transform_files | |
| − | |||
| − | |||
| − | |||
| − | : | ||
=== I don't understand your coordinate system. What do the coordinate labels R,A,S and (negative numbers) mean? === | === I don't understand your coordinate system. What do the coordinate labels R,A,S and (negative numbers) mean? === | ||
| Line 96: | Line 132: | ||
:8. ''Output Volume'':Select ''Create New Volume'' for output volume, then rename to something meaningful, like your input + suffix "_resampled" | :8. ''Output Volume'':Select ''Create New Volume'' for output volume, then rename to something meaningful, like your input + suffix "_resampled" | ||
:9. Click Apply | :9. Click Apply | ||
| + | :10. For labelmaps: go to the Volumes module and check the Labelmap box in the info tab to turn the resampled volume into a labelmap. | ||
'''Resampling in place to match another image in size''': | '''Resampling in place to match another image in size''': | ||
| − | :1. Go to the [[Documentation/{{documentation/version}}/Modules/ResampleScalarVectorDWIVolume | + | :1. Go to the [[Documentation/{{documentation/version}}/Modules/ResampleScalarVectorDWIVolume|''ResampleScalarVectorDWIVolume'' module]] |
| − | |''ResampleScalarVectorDWIVolume'' module]] | ||
:2. ''Input Volume'': Select the image you wish to resample | :2. ''Input Volume'': Select the image you wish to resample | ||
:3. ''Output Volume'': Select ''Create New Volume'' for output volume, and rename to something meaningful, like your input + suffix "_resampled" | :3. ''Output Volume'': Select ''Create New Volume'' for output volume, and rename to something meaningful, like your input + suffix "_resampled" | ||
| Line 104: | Line 140: | ||
:5. ''Interpolation Type'': check the box most appropriate for your input data: for labelmaps check ''nn=nearest Neighbor'', for 3D MRI or other bandlimited signals check ''ws=windowed sinc''. For most others leave the ''linear'' default. The ws and bspline (cubic) interpolators (''hamming, cosine, welch'') tend to produce less blurring than ''linear', but may cause overshoot near high contrast edges (e.g. negative intensity values for background pixels) | :5. ''Interpolation Type'': check the box most appropriate for your input data: for labelmaps check ''nn=nearest Neighbor'', for 3D MRI or other bandlimited signals check ''ws=windowed sinc''. For most others leave the ''linear'' default. The ws and bspline (cubic) interpolators (''hamming, cosine, welch'') tend to produce less blurring than ''linear', but may cause overshoot near high contrast edges (e.g. negative intensity values for background pixels) | ||
:6. Click Apply. Note that if the input and reference volume do not overlap in physical space, i.e. are roughly co-registered, the resampled result may not contain any or all of the input image. This is because the program will resample in the space defined by the reference image and will fill in with zeros if there is nothing at that location. If you get an empty or clipped result, that is most likely the cause. In that case try to re-center the two volumes before resampling. | :6. Click Apply. Note that if the input and reference volume do not overlap in physical space, i.e. are roughly co-registered, the resampled result may not contain any or all of the input image. This is because the program will resample in the space defined by the reference image and will fill in with zeros if there is nothing at that location. If you get an empty or clipped result, that is most likely the cause. In that case try to re-center the two volumes before resampling. | ||
| + | :7. For labelmaps: go to the Volumes module and check the Labelmap box in the info tab to turn the resampled volume into a labelmap. | ||
'''Resampling in place by specifying new dimensions''': | '''Resampling in place by specifying new dimensions''': | ||
:1. Go to the [[Documentation/{{documentation/version}}/Modules/ResampleScalarVectorDWIVolume|''ResampleScalarVectorDWIVolume'' module]] | :1. Go to the [[Documentation/{{documentation/version}}/Modules/ResampleScalarVectorDWIVolume|''ResampleScalarVectorDWIVolume'' module]] | ||
| Line 115: | Line 152: | ||
::*new image dimensions: enter new dimensions under ''Size''. To prevent clipping, the output field of view FOV = voxel size * image dimensions, should match the input | ::*new image dimensions: enter new dimensions under ''Size''. To prevent clipping, the output field of view FOV = voxel size * image dimensions, should match the input | ||
:7. leave rest at default and click ''Apply'' | :7. leave rest at default and click ''Apply'' | ||
| − | + | :8. For labelmaps: go to the Volumes module and check the Labelmap box in the info tab to turn the resampled volume into a labelmap. | |
| − | |||
== Errors == | == Errors == | ||
| Line 239: | Line 275: | ||
=== How do I generate a mask and what should it look like ? === | === How do I generate a mask and what should it look like ? === | ||
The purpose of masking (see above) is to exclude any parts of the image that are present in only one of the images yet large enough to distract the registration algorithm as it is trying to find a match for something that doesn't exist. So your mask should focus on excluding those (irrelevant) aspects of the image. Also if you have deformations in the image and care about accuracy in one region more than another you would center the mask around that region. There is a set of screencasts dedicated to quick mask building here: | The purpose of masking (see above) is to exclude any parts of the image that are present in only one of the images yet large enough to distract the registration algorithm as it is trying to find a match for something that doesn't exist. So your mask should focus on excluding those (irrelevant) aspects of the image. Also if you have deformations in the image and care about accuracy in one region more than another you would center the mask around that region. There is a set of screencasts dedicated to quick mask building here: | ||
| − | + | [[Documentation/Nightly/RegistrationVideoTutorials|Registration Video Tutorials]] | |
<br> | <br> | ||
Basically you can use any segmentation tool in Slicer that produces a labelmap. | Basically you can use any segmentation tool in Slicer that produces a labelmap. | ||
| Line 249: | Line 285: | ||
* you need a mask for both the fixed and the moving image. | * you need a mask for both the fixed and the moving image. | ||
| − | There are many example cases in the [ | + | There are many example cases in the [[Documentation:Nightly:Registration:RegistrationLibrary|'''Slicer Registration Library''']] that demonstrate the use of masks, e.g.: |
| − | *[ | + | *[[Documentation:Nightly:Registration:RegistrationLibrary:RegLib_C06|Case 06 (Breast MRI)]] |
| − | *[ | + | *[[Documentation:Nightly:Registration:RegistrationLibrary:RegLib_C07|Case 07 (prostate MRI)]] |
| − | *[ | + | *[[Documentation:Nightly:Registration:RegistrationLibrary:RegLib_C12|Case 12 (abdominal CT)]] |
| − | *[ | + | *[[Documentation:Nightly:Registration:RegistrationLibrary:RegLib_C17|Case 17 (interventional MRI-CT)]] |
=== Which registration methods offer masking? === | === Which registration methods offer masking? === | ||
| Line 263: | Line 299: | ||
=== Is there a function to convert a box ROI into a volume labelmap? === | === Is there a function to convert a box ROI into a volume labelmap? === | ||
| + | |||
| + | The [[Documentation/{{documentation/version}}/Extensions/VolumeClip ''Volume clip with ROI box'' module]] can fill a volume inside a region and outside the region with two different values. | ||
| + | |||
Slicer version 3.6 supports conversion of a box ROI into a labelmap via the [[Documentation/{{documentation/version}}/Modules/CropVolume|''Crop Volume'' module]]. This function has not (yet) been ported to Slicer4. You can create a new ROI box or select an existing one. You must select an image volume to crop for the operation, even if you're only interested in the ROI labelmap. You need not select a dedicated output for the labelmap, it is generated automatically when the cropped volume is produced, and will be called ''Subvolume_ROI_Label'' in the MRML tree. After creating the box ROI labelmap, simply delete the cropped volume and other output like the "''...resample-scale-1.0"'' volume.<br> | Slicer version 3.6 supports conversion of a box ROI into a labelmap via the [[Documentation/{{documentation/version}}/Modules/CropVolume|''Crop Volume'' module]]. This function has not (yet) been ported to Slicer4. You can create a new ROI box or select an existing one. You must select an image volume to crop for the operation, even if you're only interested in the ROI labelmap. You need not select a dedicated output for the labelmap, it is generated automatically when the cropped volume is produced, and will be called ''Subvolume_ROI_Label'' in the MRML tree. After creating the box ROI labelmap, simply delete the cropped volume and other output like the "''...resample-scale-1.0"'' volume.<br> | ||
Likely you will need the volume with the same dimension and pixel spacing as the reference image. The box volume produced above has the correct dimension, but is only 1 voxel in size. Hence there is a second step required, which is to resample the ''Subvolume_ROI_Label'' to the same resolution: use the [[Documentation/{{documentation/version}}/Modules/ResampleScalarVectorDWIVolume|''ResampleScalarVectorDWIVolume'' module]] and select the appropriate reference and ''Nearest Neighbor'' as interpolation method. Finally go to the ''Volumes'' module and check the ''Labelmap'' box in the info tab to turn the volume into a labelmap. | Likely you will need the volume with the same dimension and pixel spacing as the reference image. The box volume produced above has the correct dimension, but is only 1 voxel in size. Hence there is a second step required, which is to resample the ''Subvolume_ROI_Label'' to the same resolution: use the [[Documentation/{{documentation/version}}/Modules/ResampleScalarVectorDWIVolume|''ResampleScalarVectorDWIVolume'' module]] and select the appropriate reference and ''Nearest Neighbor'' as interpolation method. Finally go to the ''Volumes'' module and check the ''Labelmap'' box in the info tab to turn the volume into a labelmap. | ||
| Line 270: | Line 309: | ||
For further speed increase (at the cost of losing more details), you may contour just every 3rd or 4th slice and then run Dilate multiple times (until all the holes are filled in) and then run Erode as many times as you ran Dilate. You can also subsample your image first, then edit, and finally upsample again and use the SmoothLabelmap module to reduce artifacts. See FAQ below for examples on quick segmentation methods. | For further speed increase (at the cost of losing more details), you may contour just every 3rd or 4th slice and then run Dilate multiple times (until all the holes are filled in) and then run Erode as many times as you ran Dilate. You can also subsample your image first, then edit, and finally upsample again and use the SmoothLabelmap module to reduce artifacts. See FAQ below for examples on quick segmentation methods. | ||
| − | == How can I quickly generate a mask image ? == | + | === How can I quickly generate a mask image ? === |
Several tools exist to interactively segment structures. A collection of short video examples demonstrating the methods can be found here : [[Documentation/{{documentation/version}}/RegistrationVideoTutorials#Registration_Masking:_How_do_I_quickly_generate_a_mask_for_use_in_registration.3F|'''Video Tutorials on Mask generation''']] | Several tools exist to interactively segment structures. A collection of short video examples demonstrating the methods can be found here : [[Documentation/{{documentation/version}}/RegistrationVideoTutorials#Registration_Masking:_How_do_I_quickly_generate_a_mask_for_use_in_registration.3F|'''Video Tutorials on Mask generation''']] | ||
| Line 408: | Line 447: | ||
[[Image:RegLib SamplePointsBrain.png|600px|sample point densities on one slice of a brain MRI with 256x256x130 voxels; for robust registration, we recommend a useful coverage requires at least 1% coverage for affine, more (at least 5%) for nonrigid (BSpline) as DOF increase]] | [[Image:RegLib SamplePointsBrain.png|600px|sample point densities on one slice of a brain MRI with 256x256x130 voxels; for robust registration, we recommend a useful coverage requires at least 1% coverage for affine, more (at least 5%) for nonrigid (BSpline) as DOF increase]] | ||
| − | === What's the BSpline Grid Size? === | + | === Can I register 2D images with Slicer? === |
| + | Slicer is designed to work with 3D images. Since it operates in physical space, all objects must have a physical dimension (e.g. thickness). | ||
| + | It is possible to still run a 2D registration with the 3D tools, with limitations. The two options are: <br> | ||
| + | '''1.manual registration:''' the manual registration can easily be restricted to 2D. Simply do not use the sliders that control out-of plane rotation or translation. For more info on manual registration see [[Documentation/Nightly/FAQ#Can_I_manually_adjust_or_correct_a_registration.3F|'''FAQ on manual registration''']] and also [[Documentation/Nightly/Registration/RegistrationLibrary|the '''Registration Library''']] | ||
| + | <br> | ||
| + | '''2.Automated registration:''' the basic concept here is to pad the image with duplicated slices in the 3rd dimension to provide the appearance of a 3D object (e.g. using the Resample module), and then try to lock all degrees of freedom pertaining to the 3rd dimension (e.g. by masking), i.e. out-of-plane rotation or translation. Because the amount of data is very limited in 2D, automated registration is likely going to fail if the images are too far apart or to dissimilar in content. | ||
| + | An example of 2D registration can be found in our case library: | ||
| + | https://www.slicer.org/wiki/Documentation:Nightly:Registration:RegistrationLibrary:RegLib_C46 === What's the BSpline Grid Size? === | ||
The BSpline grid is the basic structure of the deformation model applied when transforming one image to match the other. A uniform grid is placed over the entire image and the grid-points are allowed to move freely. These node displacements are then interpolated over the remainder of the grid. The image below shows an example of a 3x3x3 grid. An additional row of grid points is created at the boundary of the image (red). Those points are locked and do not move. They provide a boundary constraint to keep the grid from shifting. The interior points (green) are free to move in any direction (blue arrows), which gives 3 degrees of freedom (DOF) per grid-point. So a 3x3x3 grid translates into a transform with 27 DOF, a 5x5x5 grid will have 125 DOF. For comparison a rigid transform has 6 DOF, an affine transform has 12. | The BSpline grid is the basic structure of the deformation model applied when transforming one image to match the other. A uniform grid is placed over the entire image and the grid-points are allowed to move freely. These node displacements are then interpolated over the remainder of the grid. The image below shows an example of a 3x3x3 grid. An additional row of grid points is created at the boundary of the image (red). Those points are locked and do not move. They provide a boundary constraint to keep the grid from shifting. The interior points (green) are free to move in any direction (blue arrows), which gives 3 degrees of freedom (DOF) per grid-point. So a 3x3x3 grid translates into a transform with 27 DOF, a 5x5x5 grid will have 125 DOF. For comparison a rigid transform has 6 DOF, an affine transform has 12. | ||
When specifying a grid, be aware of your image resolution and anisotropy. For example if your image has substantially fewer voxels in one direction, it may be wise to also request fewer grid-points in that dimension, to avoid over fitting.<br> | When specifying a grid, be aware of your image resolution and anisotropy. For example if your image has substantially fewer voxels in one direction, it may be wise to also request fewer grid-points in that dimension, to avoid over fitting.<br> | ||
| Line 447: | Line 493: | ||
=== How do I register images via landmarks/fiducials? === | === How do I register images via landmarks/fiducials? === | ||
| − | There are two fiducial registration tools available within Slicer. The default is within the Registration menu, and an alternative version is found in the SlicerIGT extension as "Fiducial Registration Wizard". A detailed case example using both tools is available in the [ | + | There are two fiducial registration tools available within Slicer. The default is within the Registration menu, and an alternative version is found in the SlicerIGT extension as "Fiducial Registration Wizard". A detailed case example using both tools is available in the [[Documentation:Nightly:Registration:RegistrationLibrary:RegLib_C48|Case 48 in the Registration Case Library]]. A brief screencast on selecting landmarks and running either tools are also here: |
#[[Media:RegLib_C48_FiducialRegistration.mov|'''fiducial-based registration''']] | #[[Media:RegLib_C48_FiducialRegistration.mov|'''fiducial-based registration''']] | ||
#[[Media:RegLib_C48_FiducialRegistration_IGT.mov|'''alternative tool: using SlicerIGT's fiducial-based registration''']] (shows extension download and module execution only, for the landmark selection see movie above) | #[[Media:RegLib_C48_FiducialRegistration_IGT.mov|'''alternative tool: using SlicerIGT's fiducial-based registration''']] (shows extension download and module execution only, for the landmark selection see movie above) | ||
| Line 463: | Line 509: | ||
Yes, you can nest multiple (affine) registrations inside eachother. You can generate combined ones via the right-click context menu in the ''Data'' module and selecting ''Harden Transform''. Note that the original transform or volume is replaced when selecting ''Harden Transform'', so it is recommended to rename the node afterwards to document the fact. <br> | Yes, you can nest multiple (affine) registrations inside eachother. You can generate combined ones via the right-click context menu in the ''Data'' module and selecting ''Harden Transform''. Note that the original transform or volume is replaced when selecting ''Harden Transform'', so it is recommended to rename the node afterwards to document the fact. <br> | ||
Currently (v.4.1) BSpline transforms cannot be combined with other transforms, so if you have combinations of Affine and BSpline or multiple BSpline transforms you need to resample multiple times to apply them all. We recommend to supersample the volume beforehand to counteract interpolation blurring. | Currently (v.4.1) BSpline transforms cannot be combined with other transforms, so if you have combinations of Affine and BSpline or multiple BSpline transforms you need to resample multiple times to apply them all. We recommend to supersample the volume beforehand to counteract interpolation blurring. | ||
| + | |||
| + | === Is there a module for surface registration? === | ||
| + | The current (4.5) Slicer core does not include a surface registration, but available in extensions. | ||
| + | * Rigid, similarity or affine registration of model to model: SlicerIGT extension, Model registration module (in IGT category) | ||
| + | * Rigid, similarity or affine registration of markups fiducials to model: SlicerIGT extension, Fiducials-Model registration module (in IGT category) | ||
=== Can I combine image and surface registration? === | === Can I combine image and surface registration? === | ||
| Line 506: | Line 557: | ||
Slice views: Slicer sets the overall field of view based on the image selected as "background". If the image in the foreground extends beyond that region it will be clipped. You can easily fix/test that by switching foreground and background volumes. | Slice views: Slicer sets the overall field of view based on the image selected as "background". If the image in the foreground extends beyond that region it will be clipped. You can easily fix/test that by switching foreground and background volumes. | ||
If you already created a resampled version of the registered image, that may also have been cropped,because it sets the field of view based on the reference image (which is the fixed image usually). If you place your resampled image in the background and still see cropped edges, then that's what happened. In that case either use a different reference image for resampling or no reference at all and specify the result sizes manually, or use the crop tool to expand the FOV as described below: | If you already created a resampled version of the registered image, that may also have been cropped,because it sets the field of view based on the reference image (which is the fixed image usually). If you place your resampled image in the background and still see cropped edges, then that's what happened. In that case either use a different reference image for resampling or no reference at all and specify the result sizes manually, or use the crop tool to expand the FOV as described below: | ||
| − | #use the [ | + | #use the [[Documentation/Nightly/Modules/Crop_Volume|CropVolume module]] to over-crop the reference image (define ROI to cover the field of view you want to have in the resampled volume). You will need to use interpolated crop mode, so the resolution will not match the original reference volume. |
| − | #Use the result of the cropping operation as the reference for [ | + | #Use the result of the cropping operation as the reference for [[Documentation/Nightly/Modules/BRAINSResample|BRAINSResample module]] (you will need to specify the moving volume, output volume, and the transform produced by the BRAINS registration module). |
| Line 517: | Line 568: | ||
Another alternative is to convert the transform into a 4-D deformation field directly and visualize it in slicer using RGB color. [[Documentation/{{documentation/version}}/FAQ#How_can_I_convert_a_BSpline_transform_into_a_deformation_field.3F|See FAQ below on how to convert.]] | Another alternative is to convert the transform into a 4-D deformation field directly and visualize it in slicer using RGB color. [[Documentation/{{documentation/version}}/FAQ#How_can_I_convert_a_BSpline_transform_into_a_deformation_field.3F|See FAQ below on how to convert.]] | ||
| + | === How to export the displacement magnitude of the transform as a volume? === | ||
| + | This is possible but requires use of the interactive python console: | ||
| + | transformNode=slicer.util.getNode('LinearTransform_3') | ||
| + | referenceVolumeNode=slicer.util.getNode('MRHead') | ||
| + | slicer.modules.transforms.logic().CreateDisplacementVolumeFromTransform(transformNode, referenceVolumeNode, False) | ||
| + | The new volume can then be saved or used for ROI analysis with the [[Documentation/Nightly/Modules/LabelStatistics|Label Statistics module]]. | ||
=== Physical Space vs. Image Space: how do I align two registered images to the same image grid? === | === Physical Space vs. Image Space: how do I align two registered images to the same image grid? === | ||
| − | Slicer displays all data in a physical coordinate system. Hence an image can only be displayed correctly if it contains sufficient header information to relate the image voxel grid with physical space. This includes voxel size, axis orientation and scan order. It is therefore possible for two images to be aligned when viewed in Slicer, even though their underlying image grid is oriented very differently. To match the two images in image as well as physical space, the abovementioned axis direction, voxel size and image grid orientation must match. The procedure will depend on the image data, but the main tools at your disposal are the [[Documentation/{{documentation/version}}/Modules/ResampleScalarVectorDWIVolume|''ResampleScalarVectorDWIVolume'' module]] and [[Documentation/{{documentation/version}}/Modules/OrientImages|''OrientImages'' module]]. | + | Slicer displays all data in a physical coordinate system. Hence an image can only be displayed correctly if it contains sufficient header information to relate the image voxel grid with physical space. This includes voxel size, axis orientation and scan order. It is therefore possible for two images to be aligned when viewed in Slicer, even though their underlying image grid is oriented very differently. To match the two images in image as well as physical space, the abovementioned axis direction, voxel size and image grid orientation must match. The procedure will depend on the image data, but the main tools at your disposal are the [[Documentation/{{documentation/version}}/Modules/ResampleScalarVectorDWIVolume|''ResampleScalarVectorDWIVolume'' module]] and [[Documentation/{{documentation/version}}/Modules/OrientImages|''OrientImages'' module]]. |
| − | |||
== Nonrigid / BSpline Registration == | == Nonrigid / BSpline Registration == | ||
| Line 534: | Line 590: | ||
=== How can I convert a BSpline transform into a deformation field? === | === How can I convert a BSpline transform into a deformation field? === | ||
| − | There is | + | There is a dedicated module to convert a BSpline ITK transform file (.tfm) into a deformation field volume. <br> |
| + | Modules -> all Modules -> '''BSpline to deformation field'''<br> | ||
| + | provide your BSpline transform and a reference image that defines the size of the deformation field. | ||
| + | <br>For Slicer 3.6 this functionality was available as command line functionality only: | ||
| + | To execute, type (exchange ''/Applications/Slicer3.6.3'' with the path of your Slicer installation): | ||
/Applications/Slicer3.6.3/Slicer3 --launch /Applications/Slicer3.6.3/lib/Slicer3/Plugins/BSplineToDeformationField --tfm InputBSpline.tfm | /Applications/Slicer3.6.3/Slicer3 --launch /Applications/Slicer3.6.3/lib/Slicer3/Plugins/BSplineToDeformationField --tfm InputBSpline.tfm | ||
--refImage ReferenceImage.nrrd --defImage Output_DeformationField.nrrd | --refImage ReferenceImage.nrrd --defImage Output_DeformationField.nrrd | ||
for more details try: | for more details try: | ||
/Applications/Slicer3.6.3/Slicer3 --launch /Applications/Slicer3.6.3/lib/Slicer3/Plugins/BSplineToDeformationField --help | /Applications/Slicer3.6.3/Slicer3 --launch /Applications/Slicer3.6.3/lib/Slicer3/Plugins/BSplineToDeformationField --help | ||
| + | |||
| + | === Where can I check the reference color map for the transform visualizer? i.e. how can I know the magnitude of transform vectors? === | ||
| + | The "Transform Visualizer" is a tool that in earlier Slicer versions (4.2 and before) was an extension module, but in newer versions has been integrated into the [[Documentation/{{documentation/version}}/Modules/Transforms|'''Transforms''' module]]] (under the ''Display'' tab). It enables visualisation of transform vectors that are particularly useful to help understand complex non-rigid deformations such as B-Spline registrations. | ||
| + | You can find the referenced color table in the [[Documentation/{{documentation/version}}/Modules/Colors|'''Colors''' module]] . There is a category called “Transform Display” and under that there is the “Displacement to color” color table. To display the scalar value represented by a color, you need to turn on the “Display scalar bar” option in Scalar Bar section under “Colors” module. Currently it only shows the scalar bar in the 3D window. <br> | ||
| + | Alternatlively, you can see the deformation vector under current cursor in Transform module in the Information section, when you move your cursor, it will display the current vector. | ||
Latest revision as of 13:20, 27 November 2019
Home < Documentation < Nightly < FAQ < Registration
|
For the latest Slicer documentation, visit the read-the-docs. |
Contents
- 1 Registration
- 1.1 Spatial Orientation, Header, Image Size
- 1.1.1 How do I fix incorrect axis directions? Can I flip an image (left/right, anterior/posterior etc) ?
- 1.1.2 How do I fix a wrong image orientation in the header? / My image appears upside down / facing the wrong way / I have incorrect/missing axis orientation
- 1.1.3 Can I undo the "centering" of an image
- 1.1.4 I have some DICOM images that I want to reslice at an arbitrary angle
- 1.1.5 How do I fix incorrect voxel size / aspect ratio of a loaded image volume?
- 1.1.6 The registration transform file saved by Slicer does not seem to match what is shown
- 1.1.7 I don't understand your coordinate system. What do the coordinate labels R,A,S and (negative numbers) mean?
- 1.1.8 My image is very large, how do I downsample to a smaller size?
- 1.2 Errors
- 1.2.1 Registration failed with an error. What should I try next?
- 1.2.2 Registration result is wrong or worse than before?
- 1.2.3 Registration results are inconsistent and don't work on some image pairs. Are there ways to make registration more robust?
- 1.2.4 How do I register images that are very far apart / do not overlap
- 1.2.5 How do I initialize/align images with very different orientations and no overlap?
- 1.2.6 Can I manually adjust or correct a registration?
- 1.3 Diffusion
- 1.4 Masking
- 1.4.1 What's the purpose of masking / VOI in registration? / What does the masking option in registration accomplish ?
- 1.4.2 How do I generate a mask and what should it look like ?
- 1.4.3 Which registration methods offer masking?
- 1.4.4 Is there a function to convert a box ROI into a volume labelmap?
- 1.4.5 I have to manually segment a large number of slices. How can I make the process faster?
- 1.4.6 How can I quickly generate a mask image ?
- 1.5 Parameters & Concept
- 1.5.1 What's the difference between Rigid and Affine registration?
- 1.5.2 What's the difference between Affine and BSpline registration?
- 1.5.3 What's the difference between the various registration methods listed in Slicer?
- 1.5.4 How many sample points should I choose for my registration?
- 1.5.5 Can I register 2D images with Slicer?
- 1.5.6 I want to register two images with different intensity/contrast.
- 1.5.7 How important is bias field correction / intensity inhomogeneity correction?
- 1.5.8 Have the Slicer registration methods been validated?
- 1.5.9 How can I save the parameter settings I have selected for later use or sharing?
- 1.5.10 Registration is too slow. How can I speed up my registration?
- 1.5.11 How do I register images via landmarks/fiducials?
- 1.5.12 Is the BRAINSfit registration for brain images only?
- 1.5.13 One of my images has a clipped field of view. Can I still use automated registration?
- 1.5.14 Can I combine multiple registrations?
- 1.5.15 Is there a module for surface registration?
- 1.5.16 Can I combine image and surface registration?
- 1.6 Apply, Resample, Export
- 1.6.1 I ran a registration but cannot see the result. How do I visualize the result transform?
- 1.6.2 My reoriented image returns to original position when saved; Problem with the Harden Transform function
- 1.6.3 What is the Meaning of 'Fixed Parameters' in the transform file (.tfm) of a BSpline registration ?
- 1.6.4 After registration the registered image appears cropped. How can I increase the field of view to see/include the entire image
- 1.6.5 How can I see the parameters of the function that describe a BSpline registration/deformation?
- 1.6.6 How to export the displacement magnitude of the transform as a volume?
- 1.6.7 Physical Space vs. Image Space: how do I align two registered images to the same image grid?
- 1.7 Nonrigid / BSpline Registration
- 1.7.1 The nonrigid (BSpline) registration transform does not seem to be nonrigid or does not show up correctly.
- 1.7.2 What's the difference between BRAINSfit and BRAINSDemonWarp?
- 1.7.3 How can I convert a BSpline transform into a deformation field?
- 1.7.4 Where can I check the reference color map for the transform visualizer? i.e. how can I know the magnitude of transform vectors?
- 1.1 Spatial Orientation, Header, Image Size
Registration
Spatial Orientation, Header, Image Size
How do I fix incorrect axis directions? Can I flip an image (left/right, anterior/posterior etc) ?
Sometimes the header information that describes the orientation and size of the image in physical space is incorrect or missing. Slicer displays images in physical space, in a RAS orientation. If images appear flipped or upside down, the transform that describes how the image grid relates to the physical world is incorrect. In proper RAS orientation, a head should have anterior end at the top in the axial view, look to the left in a sagittal view, and have the superior end at the top in sagittal and coronal views.
Yes, you can flip images and change the axis orientation of images in slicer. But we urge to use great caution when doing so, since this can introduce subtantial problems if done wrong. Worse than no information is wrong information. Below the steps to flip the LR axis of an image:
You may skip steps 5-8 below and download predefined transforms here. To apply those unzip, drag & drop into Slicer and drag your volume inside the transform.
- Go to the Data module, right click on the node labeled "Scene" and select "Insert Transform" from the pulldown menu
- You should see a new transform node being added to the tree, named "LinearTransform_1" or similar.
- left click on the volume you wish to flip, and drag it onto the new transform node. You should see a "+" appear in front of the transform node, and clicking on it should reveal the volume now inside/under that transform.
- make sure you have the image you wish to flip selected and visible in the slice views, preferably all 3 views (sagittal, coronal, axial).
- Switch to the Transforms module and (if not selected already) select the newly created transform from the Active Transform menu.
- Under Transform Matrix you see a 4x4 array of ones and zeros. Each row represents an axis direction. We will switch the axis direction by changing the sign of one of the 1s.
- e.g. to flip left/right: double-click inside the top left field where you see a 1. The number should be high-lighted and change to 1.0
- replace the 1.0 with "-1.0", then hit the RETURN key. You should see a flip immediately, assuming you have the volume in the proper view. Depending on which of the axes you want flipped, select the 1 in one of the other rows.
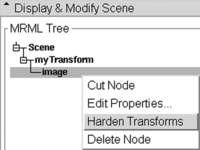
- When you have the result you want, return to the Data module, there right click on your image and select Harden Transform from the pulldown menu.
- The image will move back outside the transform onto the main level, indicating that your change of axis orientation has now been applied. Note there is no Undo for this step. If you change your mind you have to apply the same flip again or reload your volume.
- Note that the flip will likely also cause a shift, depending on your image origin. You may choose to recenter your image. To do so go to the Volumes module, open the Volume Information tab and click on the Center Volume button
- Note that this change was applied to the header information that stores the physical orientation, not the image data itself. Hence you will only see this flip in software that reads and accounts this header orientation info.
- Save your image under a new name, do not use a format that doesn't store physical orientation info in the header (jpg, gif etc).; also consider saving the transform as documentation to what change you have applied. You can also use these saved transforms as templates to quickly flip an image.
- Again: the change is saved as part of the image orientation info and not as an actual resampling of the image, i.e. if you save your image and reload it in another software that does not read the image orientation info in the header (or displays in image space only), you will not see the change you just applied.
To flip the other axes do the same as above but edit the diagonal entries in the 2nd and 3rd row, for flipping anterior-posterior and inferior-superior directions, respectively.
How do I fix a wrong image orientation in the header? / My image appears upside down / facing the wrong way / I have incorrect/missing axis orientation
- Problem: My image appears upside down / flipped / facing the wrong way / I have incorrect/missing axis orientation
- Explanation: Slicer presents and interacts with images in physical space, which differs from the way the image is stored by a separate transform that defines how large the voxels are and how the image is oriented in space, e.g. which side is left or right. This information is stored in the image header, and different image file formats have different ways of storing this information. If Slicer supports the image format, it should read the information in the header and display the image correctly. If the image appears upside down or with distorted aspect ratio etc, then the image header information is either missing or incorrect.
- Fix: See the FAQ above on how to flip an image axis within Slicer. You can also correct the voxel dimensions and the image origin in the Volume Information tab of the Volumes module, and you can reorient images via the Transforms module.
- To fix an axis orientation directly in the header info of an image file:
- 1. load the image into slicer (File: Add Volume, Add Data, Load Scene..)
- 2. save the image back out as NRRD (.nhdr) format.
- 3. open the .nhdr with a text editor of your choice. You should see a line that looks like this:
space: left-posterior-superior sizes: 448 448 128 space directions: (0.5,0,0) (0,0.5,0) (0,0,0.8)
- 4. the three brackets ( ) represent the coordinate axes as defined in the space line above, i.e. the first one is left-right, the second anterior-posterior, and the last inferior-superior. To flip an axis place a minus sign in front of the respective number, which is the voxel dimension. E.g. to flip left-right, change the line to
space directions: (-0.5,0,0) (0,0.5,0) (0,0,0.8)
- 5. alternatively if the entire orientation is wrong, i.e. coronal slices appear in the axial view etc., you may easier just change the space field to the proper orientation. Note that Slicer uses RAS space by default, i.e. first (x) axis = left-right, second (y) axis = posterior-anterior, third (z) axis = inferior-superior
- 6. save & close the edited .nhdr file and reload the image in slicer to see if the orientation is now correct.
Can I undo the "centering" of an image
When importing images, there's a "Center" checkbox, which if checked will reset the image origin to the center of the image grid, and ignore the image origin stored in the header. The same function is available to loaded images in the Volumes module (Volume Information Tab). Results derived from images have their spatial info stored relative to that image origin. So fiducial points or label maps obtained from centered images will also be centered, which means they will align with a centered version of the image but not the original one. Is there a way to return such data to the original uncentered position?
There is no dedicated module or function for that purpose currently implemented, but there are several ways to return data to the position before centering, provided the original image with the old origin is still available. Options are:
- copy the image origin (or entire spatial orientation info) from the original reference image into the header of the "centered" image. For images stored in a format where the header data is accessible in text format this is fairly straightforward. For other formats with binary headers it will require dedicated software to read the header and re-save the image.
- create a transform that embodies the shift and apply it to the data. This is probably the most accessible solution. It will work for all forms of data, i.e. fiducial points, labelmaps, surface models etc. To manually obtain such a transform:
- Load both original and centered image
- Go to the Volumes module, open the Volume Information Tab, then select either image and record the "Image Origin" information displayed. Calculate the difference of the two origins (x1-x2, y1-y2, z1-z2).
- Go to the Transforms module , create a new transform, then enter the origin difference calculated above into the fields for translation (LR= left-right, PA=posterior-anterios, IS=inferior-superior). Note that to replicate the effect of centring the translation vector is centered-uncentered; to go back from centered to uncentered is the inverse of that transform, i.e. uncentered-centered. The "Invert Transform" button in the transforms module lets you switch between the two.
- Go to the Data module. Drag the centered image and any data derived from it 'into the transform.
- Set fore- & background to original and centered image to verify, set the fade slider halfway so you can see both images. Verify that the two images align once the "centered" image has been placed into the transform.
- Right click on the meta data (e.g. fiducials) inside the transform, select Harden Transform from the pulldown menu. The data node will move back out of the transform to the main level to indicate the transform has been applied.
- Rename the data node (double click) to document that it has shifted. Then save it as a new file.
See here for a screencast describing this procedure to "uncenter" and shift meta data back to the original position.
For large sets of images calculating the offset manually may not be feasible. Below is a rudimentary python script that will read two or more images (NIfTI format) , calculate the offset and save it as an ITK transform (.tfm) file. You can then import this transform into Slicer and apply it to the data that needs shifting.
- As another alternative you can also run a quick automated registration of the centered to the uncentered image to obtain a transform. Note that this will look for a match based on image similarity and will not be 100% precise, but likely very close.
#! /usr/bin/env python # reads 2 or more NIfTI images and extracts the image center offset to the first as an ITK transform file v1.0 # usage: NIICenterOffset2ITK.py RefImg.nii CenteredImg1.nii CenteredImg2.nii ... # output: CenteredImg1_center.tfm CenteredImg2_center.tfm ...
import nibabel as nib
import numpy
import sys
refimg = nib.load(sys.argv[1])
refhdr = refimg.get_header()
reforigin=refhdr.get_qform()[([0,1,2],3)].astype(float)
for aImgName in sys.argv[2:]:
img = nib.load(aImgName)
hdr = img.get_header()
origin=hdr.get_qform()[([0,1,2],3)].astype(float)
offset=reforigin-origin
itk_file = open(aImgName+'.centered.tfm', "w")
itk_file.write('#Insight Transform File V1.0\n#Transform 0\nTransform: AffineTransform_double_3_3\n' )
itk_file.write('Parameters: 1 0 0 0 1 0 0 0 1 %f %f %f\n' %( tuple( (reforigin-origin).tolist())) )
itk_file.write('FixedParameters: 0 0 0\n')
itk_file.close()
I have some DICOM images that I want to reslice at an arbitrary angle
There's several ways to go about this. If you wish to register your image to another reference/target image, run one of the automated registration methods. If you wish to realign manually, most efficient way is to use the Transforms module. Once you have the desired orientation, you need to apply the new orientation to the image. You can do this in 2 ways: 1) without or 2) with resampling the image data.
- Without resampling: In the Data module, select the image (inside the transforms node) and select "Harden Transforms" from the pulldown menu. This will write the new orientation in physical space into the image header. This will work only if other software you use and the image format you save it as support this form of orientation information in the image header.
- With resampling: Go to the ResampleScalarVectorDWIVolume module and create a new image by resampling the original with the new transform. This will incur interpolation blurring but is guaranteed to transfer for all image formats or software.
For more details on manual transform, see this FAQ and the here.
How do I fix incorrect voxel size / aspect ratio of a loaded image volume?
- Problem: My image appears distorted / stretched / with incorrect aspect ratio
- Explanation: Slicer presents and interacts with images in physical space, which differs from the way the image is stored by a set of separate information that represents the physical "voxel size" and the direction/spatial orientation of the axes. If the voxel dimensions are incorrect or missing, the image will be displayed in a distorted fashion. This information is stored in the image header. If the information is missing, a default of isotropic 1 x 1 x 1 mm size is assumed for the voxel.
- Fix: You can correct the voxel dimensions and the image origin in the Info tab of the Volumes module. If you know the correct voxel size, enter it in the fields provided (double click to edit). You should see the display update immediately. Ideally you should try to maintain the original image header information from the point of acquisition. Sometimes this information is lost in format conversion. Try an alternative converter or image format if you know that the voxel size is correctly stored in the original image. Alternatively you can try to edit the information in the image header, e.g. save the volume as (NRRD (.nhdr) format and open the ".nhdr" file with a text editor. See FAQ above.
The registration transform file saved by Slicer does not seem to match what is shown
When executing the following procedure:
- Create a transform.
- Adjust it by adjusting the 6 slider bars in the Transforms module.
- Save the transform as a .tfm file.
- Inspect the contents of the .tfm file in a text editor, and compare them to what is shown in the 4x4 matrix in the Transforms module.
- re-load the .tfm back into slicer and confirm you have the same data as you saved from Slicer.
You will notice that the original and re-loaded Transforms are identical, but do not match the content of the .tfm file. The issue relates to the difference between Slicer, which uses a "computer graphics" view of the world, and ITK, which uses an "image processing" view of the world. The Slicer transform hierarchy models movement of an object from one spot to another. For example, a transform that has a positive "superior" value wrapped around a volume moves the volume up in patient space.
Conversely, ITK transforms map "backwards": from the display space back to the original image. Imagine stepping sequentially through the output pixels: ITK wants to know the transform back to the input pixels used to calculate the output. Additionally ITK transforms are saved in LPS, whereas the Slicer Transform widget uses RAS coordinates.
In Summary:
- The transform represented in the widget is in RAS.
- The transform represented in the tfm file is in LPS.
- The transform represented in the file is the inverse of the transform in the widget, and has the LPS/RAS conversion applied.
- The order of the parameters in the tfm are the elements of the upper 3x3 of the transform displayed in the widget followed by the elements in the last column of the widget.
Please see this discussion for more information, and code examples:
https://www.slicer.org/wiki/Documentation/Nightly/Modules/Transforms#Transform_files
I don't understand your coordinate system. What do the coordinate labels R,A,S and (negative numbers) mean?
- It's very important to realize that Slicer displays all images in physical space, i.e. in mm. This requires orientation and size information that is stored in the image header. How that header info is set and read from the header will determine how the image appears in Slicer. RAS is the abbreviation for right, anterior, superior; indicating in order the relation of the physical axis directions to how the image data is stored.
- For a detailed description on coordinate systems see here.
My image is very large, how do I downsample to a smaller size?
Several Resampling modules provide this functionality. If you also have a transform you wish to apply to the volume, we recommend the ResampleScalarVectorDWIVolume module, or the simpler ResampleScalarVolume module. See here for an explanation and overview of Resampling tools.
Resampling in place (changing voxel size):
- 1. Go to the Volumes module
- 2. from the Active Volume pulldown menu, select the image you wish to downsample
- 3. Open the Volume Information tab. Write down the voxel dimensions (Image Spacing) and overall image size (Image Dimensions), e.g. 1.2 x 1.2 x 3 mm voxel size, 512 x 512 x 86. You will need this information to determine the amount of down-/up-sampling you wish to apply
- 4. Go to the ResampleScalarVolume module (found under All modules)
- 5. In the Spacing field, enter the new desired voxel size. This is the above original voxel size multiplied with your downsampling factor. For example, if you wish to reduce the image to half (in plane), but leave the number of slices, you would enter a new voxel size of 2.4,2.4,3.
- 6. For Interpolation, check the box most appropriate for your input data: for labelmaps check nearest Neighbor, for 3D MRI or other bandlimited signals check hamming. For most others leave the linear default. The sinc interpolator (hamming, cosine, welch) and bspline (cubic) interpolators tend to produce less blurring than linear', but may cause overshoot near high contrast edges (e.g. negative intensity values for background pixels)
- 7.Input Volume: Select the image you wish to resample
- 8. Output Volume:Select Create New Volume for output volume, then rename to something meaningful, like your input + suffix "_resampled"
- 9. Click Apply
- 10. For labelmaps: go to the Volumes module and check the Labelmap box in the info tab to turn the resampled volume into a labelmap.
Resampling in place to match another image in size:
- 1. Go to the ResampleScalarVectorDWIVolume module
- 2. Input Volume: Select the image you wish to resample
- 3. Output Volume: Select Create New Volume for output volume, and rename to something meaningful, like your input + suffix "_resampled"
- 4. Reference Volume: Select the reference image whose size/dimensions you want to match to.
- 5. Interpolation Type: check the box most appropriate for your input data: for labelmaps check nn=nearest Neighbor, for 3D MRI or other bandlimited signals check ws=windowed sinc. For most others leave the linear default. The ws and bspline (cubic) interpolators (hamming, cosine, welch) tend to produce less blurring than linear', but may cause overshoot near high contrast edges (e.g. negative intensity values for background pixels)
- 6. Click Apply. Note that if the input and reference volume do not overlap in physical space, i.e. are roughly co-registered, the resampled result may not contain any or all of the input image. This is because the program will resample in the space defined by the reference image and will fill in with zeros if there is nothing at that location. If you get an empty or clipped result, that is most likely the cause. In that case try to re-center the two volumes before resampling.
- 7. For labelmaps: go to the Volumes module and check the Labelmap box in the info tab to turn the resampled volume into a labelmap.
Resampling in place by specifying new dimensions:
- 1. Go to the ResampleScalarVectorDWIVolume module
- 2. Input Volume: Select the image you wish to resample
- 3. Output Volume: Select Create New Volume for output volume, and rename to something meaningful, like your input + suffix "_resampled"
- 4. Reference Volume: leave at "none"
- 5. Interpolation Type: check the box most appropriate for your input data: for labelmaps check nn=nearest Neighbor, for 3D MRI or other bandlimited signals check ws=windowed sinc. For most others leave the linear default. The ws and bspline (cubic) interpolators (hamming, cosine, welch) tend to produce less blurring than linear', but may cause overshoot near high contrast edges (e.g. negative intensity values for background pixels)
- 6. Click Apply. Note that if the input and reference volume do not overlap in physical space, i.e. are roughly co-registered, the resampled result may not contain any or all of the input image. This is
- 6. Manual Output Parameters: here you specify the new voxel size / spacing and dimensions. Note that you need to set both. If only the voxel size is specified, the image is resampled but retains its original dimensions (i.e. empty/zero space). If only the dimensions are specified the image will be resampled starting at the origin and cropped but not resized.
- new voxel size: calculate the new voxel size and enter in the Spacing field, as described above in in 'Resampling in place above, see step #5
- new image dimensions: enter new dimensions under Size. To prevent clipping, the output field of view FOV = voxel size * image dimensions, should match the input
- 7. leave rest at default and click Apply
- 8. For labelmaps: go to the Volumes module and check the Labelmap box in the info tab to turn the resampled volume into a labelmap.
Errors
Registration failed with an error. What should I try next?
- Problem: automated registration fails, status message says "completed with error" or similar.
- Explanation: Registration methods are mostly implemented as commandline modules, where the input to the algorithm is provided as temporary files and the algorithm then seeks a solution independently from the activity of the Slicer GUI. you will notice that you're free to continue using other Slicer functions while a registration is running. Several reasons can lead to failure, most commonly they are wrong or inconsistent input or lack of convergence if images are too far apart initially.
- check your input:
- did you provide both a fixed and a moving image?
- did you select an output (transform and/or new output volume)?
- do the two images have any overlap? Can you see them both in the slice views? If not try to recenter (see "Manual Recenter" in FAQ above). When rerunning the registration, try selecting an initializer
- are inputs consistent? E.g. General Registration (BRAINS) module will complain if you check a "BSpline" registration phase but do not select a BSpline output transform, or if you request masking and do not specify masking input or output.
- if above checks reveal nothing, open the Error Log window (Window Menu) and scroll to the bottom to see the most recent entries related to the registration. Usually you will see a commandline entry that shows which arguments were given to the algorithm, and a standard output or similar that lists what the algorithm returned. More detailed error info can be found in either this entry, or in the ERROR: ..." line at the top of the list. Click on the corresponding line and look for explanation in the provided text. If there was a problem with the input arguments or the that would be reported here.
- If the Error log does not provide useful clues, try varying some of the parameters. Note that if the algorithm aborts/fails right away and returns immediately with an error, most likely some input is wrong/inconsistent or missing.
- check the initial misalignment, if images are too far apart and there is no overlap, registration may fail. Consider initialization with a prior manual alignment, centering the images or using one of the initialization methods provided by the modules
- write to the Slicer user group (slicer-users@bwh.harvard.edu) and inform them of the error. We're keen on learning so we can improve the program. The fastest and best reply you will get if you copy and paste the error messages found in the Error Log into your mail.
Registration result is wrong or worse than before?
- Problem: automated registration provides an alignment that is insufficient, possibly worse than the initial position
- Explanation: The automated registration algorithms (except for fiducial and manual registration) in Slicer operate on image intensity and try to move images so that similar image content is aligned. This is influenced by many factors such as image contrast, resolution, voxel anisotropy, artifacts such as motion or intensity inhomogeneity, pathology etc, the initial misalignment and the parameters selected for the registration.
- re-run the registration with parameter modifications:
- if images have little initial overlap or are far apart in orientation, try a (different) initializer: e.g. General Registration (BRAINS) module has several initializers, details in their documentation and also in this FAQ above.
- do the two images have any overlap? Can you see them both in the slice views? If not try to recenter (see "Manual Recenter" in FAQ above). When
- try a lower DOF registration first to see if that fails. If the lower DOF fails, subsequent ones will also. For nonrigid registration, try adding intermediate steps, such as a similarity (7 DOF) or Affine (12 DOF) transform.
- if automated initializers do not help, try a manual initial alignment (see FAQ above). This need not be perfect, as long as it establishes good overlap and roughly same direction. Then try rerunning the registration using the manual transform as a starting point. (Initialization Transform) in the General Registration (BRAINS) module.
- if initial overlap is ok but registration "drifts away", there is either insufficient sample data or distracting image content. Try increasing the number of sample points. See FAQ below for estimates of sample points.
- insufficient contrast: consider adjusting the Histogram Bins (where avail.) to tune the algorithm to weigh small intensity variations more or less heavily
- strong anisotropy:' if one or both of the images have strong voxel anisotropy of ratios 5 or more, rotational alignment may become increasingly difficult for an automated method. Consider increasing the sample points and reducing the Histogram Bins.
- distracting image content: pathology, strong edges, clipped FOV with image content at the border of the image can easily dominate the cost function driving the registration algorithm. Masking is a powerful remedy for this problem: create a mask (binary labelmap/segmentation) that excludes the distracting parts and includes only those areas of the image where matching content exists. This requires one of the modules that supports masking input, such as General Registration (BRAINS) module or Expert Automated Registration module . Next best thing to use with modules that do not support masking is to mask the image manually and create a temporary masked image where the excluded content is set to 0 intensity; the MaskScalarVolume module performs this task.
- you can adjust/correct an obtained registration manually, within limits, as outlined manual initial alignment in this FAQ above.
Registration results are inconsistent and don't work on some image pairs. Are there ways to make registration more robust?
The key parameters that influence registration robustness are the number of sample points, the initial degrees of freedom of the transform, the type of similarity metric and the initial misalignment and image contrast/content differences. particularly initialization methods that seek a first alignment before beginning the optimization can make things worse. If initial position is already sufficiently close (i.e. more than 70% overlap and less than 20% rotational misalignment), consider turning off initialization if available (e.g. in General Registration (BRAINS) module (under Registration menu) and the Expert Automated Registration module )
Try increasing the sample points. Guidelines on selecting sample points are given here. The degrees of freedom usually are given by the overall task and not subject to variation, but depending on initial misalignment, robustness can greatly improve by an iterative approach that gradually increases DOF rather than starting with a high DOF setting. The General Registration (BRAINS) module and Expert Automated Registration module both allow prescriptions of iterative DOF.
If using a cost/criterion function other than mutual information (MI), note that MI tends to be the most forgiving/robust toward differences in image contrast.
Also see the.
How do I register images that are very far apart / do not overlap
- Problem: when you place one image in the background and another in the foreground, the one in the foreground will not be visible (entirely) when switching bak & forth
- Explanation:Slicer chooses the field of view (FOV) for the display based on the image selected for the background. The FOV will therefore be centered around what is defined in that image's origin. If two images have origins that differ significantly, they cannot be viewed well simultaneously.
- Fix: recenter one or both images as follows:
- 1. Go to the Volumes module
- 2. Select the image to recenter from the Active Volume menu
- 3. Select/open the Volume Information tab.
- 4. Click the Center Volume button. You will notice how the Image Origin numbers displayed above the button change. If you have the image selected as foreground or background, you may see it move to a new location.
- 5. Repeat steps 2-4 for the other image volumes
- 6. In the slice view menu, click on the Fit to Window button (a small square next to the pin in the top left corner of each view)
- 7. Images should now be roughly in the same space. Note that this re-centering is considered a change to the image volume, and Slicer will mark the image for saving next time you select Save.
How do I initialize/align images with very different orientations and no overlap?
I would like to register two datasets, but the centers of the two images are so different that they don't overlap at all. Is there a way to pre-register them automatically or manually to create an initial starting transformation?
- Manual Recenter: See the FAQ above for re-centering images
- 1. Go to the Volumes module
- 2. Select the image to recenter from the Active Volume menu
- 3. Select/open the Volume Information tab.
- 4. Click the Center Volume button. You will notice how the Image Origin numbers displayed above the button change. If you have the image selected as foreground or background, you may see it move to a new location.
- 5. Repeat steps 2-4 for the other image volumes
- 6. In the slice view menu, click on the Fit to Window button (a small square next to the pin in the top left corner of each view)
- 7. Images should now be roughly in the same space. Note that this re-centering is considered a change to the image volume, and Slicer will mark the image for saving next time you select Save.
- Automatic Initialization: Most registration tools have initializers that should take care of the initial alignment in a scenario you described. However since they often are based on heuristics they may work well in some cases and not in others. The two modules that offer the most initializer options are General Registration (BRAINS) module (under Registration menu) and the Expert Automated Registration module.
- General Registration (BRAINS) module initializers:
- Initialization Transform: here you can specify a transform from which to start. You can perform a manual alignment (see here for tutorial) and then feed this as initializer here.
- Initialization Transform Mode: these options generate automated initializations for you:
- "Off assumes that the physical space of the images are close, and that centering in terms of the image Origins is a good starting point.
- useCenterOfHeadAlign: recommended for registering brain MRI where all or most of the head is within the FOV
- useMomentsAlign: recommended for image pairs with similar contrast, scale and content.
- useGeometryAlign: recommended for image pairs with similar FOV for both objects. This aligns the image grid volumes disregarding of content.
- useCenterOfROIAlign": recommended if you have a mask for each image that defines the regions you want registered. This will initialize based on those two masks.
- Tip: yo can run the registration with just the initializer to see what kind of transformation it produces. In that case select a Slicer Linear Transform output but leave all boxes under Registration Phases unchecked.
- ExpertAutomatedRegistration module initializers:
- "None directly starts with optimization from the current position
- CentersOfMass: recommended for image pairs with similar contrast, scale and content. Similar to useMomentsAlign above.
- SecondMoments: same as above, but also calculating (principal) axis directions
- Image Centers": similar to useGeometryAlign above: recommended for image pairs with similar FOV for both objects. This aligns the image grid volumes disregarding of content.
Can I manually adjust or correct a registration?
Yes for linear (rigid to affine) transforms; not without resampling for nonrigid transforms.
The automated registration algorithms (except for fiducial and surface registration) in Slicer operate on image intensity and try to move images so that similar image content is aligned. This is influenced by many factors such as image contrast, resolution, voxel anisotropy, artifacts such as motion or intensity inhomogeneity, pathology etc, the initial misalignment and the parameters selected for the registration. Before attempting manual correction, it is usually advisable to retry an automated run with modified parameters, additional initializers or masks.
you can adjust/correct an obtained registration manually, within limits. There's a brief (no sound) video that demonstrates the procedure here: Manual Registration Movie (2 min)
If the transform is linear, i.e. a rigid or affine transform, you can access the rigid components (translation and rotation) of that transform via the Transforms module. Or (maybe safer) you can create an additional new transform and nest the old one inside it. Then once you approve of the adjustment merge the two (via Harden Transform)
- Go to the Data module, right click on the node labeled "Scene" and select "Insert Transform" from the pulldown menu
- You should see a new transform node being added to the tree, named "LinearTransform_1" or similar.
- left click on the volume you wish to register, and drag it onto the new transform node. You should see a "+" appear in front of the transform node, and clicking on it should reveal the volume now inside/under that transform.
- make sure you have the image for which you wish to adjust the registration selected and visible in the slice views, preferably all 3 views (sagittal, coronal, axial).
- Switch to the Transforms module and (if not selected already) select the newly created transform from the Active Transform menu.
- adjust the translation and rotation sliders to adjust the current position. To get a finer degree of control, enter smaller numbers for the translation limits and enter rotation angles numerically in increments of a few degrees at a time
Diffusion
How do I register a DWI image dataset to a structural reference scan? (Cookbook)
- Problem: The DWI/DTI image is not in the same orientation as the reference image that I would like to use to locate particular anatomy; the DWI image is distorted and does not line up well with the structural images
- Explanation: DWI images are often acquired as EPI sequences that contain significant distortions, particularly in the frontal areas. Also because the image is acquired before or after the structural scans, the subject may have moved in between and the position is no longer the same.
- Fix: obtain a baseline image from the DWI sequence, register that with the structural image and then apply the obtained transform to the DTI tensor. The two chief issues with this procedure deal with the difference in image contrast between the DWI and the structural scan, and with the common anisotropy of DWI data.
- Overall Strategy and detailed instructions for registration & resampling can be found in our DWI registration cookbook
- you can find example cases in the DWI chapter of the Slicer Registration Case Library, which includes example datasets and step-by-step instructions. Find an example closest to your scenario and Slicer version and perform the registration steps recommended there.
Is there a way to perform an Eddy current correction on DWI in Slicer
There is a way, you need to download the GTRACT extension (you can get it from the menu View->Extension Manager) Once you have it you will see a new module category under the diffusion one called "GTRACT". Within this category there is a module called "Coregister B-values". This module takes a DWI image and outputs a DWI image in which every DWI is co-registered to one of the B0 images (by default the first one), this can be regarded as motion correction. Within this module there is a checkbox in the "Registration Parameters" section: "Eddy Current Correction". This will tune some of the registration parameters such that some Eddy Current artifacts are corrected.
Masking
What's the purpose of masking / VOI in registration? / What does the masking option in registration accomplish ?
The masking option is a very effective tool to focus the registration onto the image content that is most important. It is often the case that the alignment of the two images is more important in some areas than others. Automated registration based on image intensity is easily dominated by the portions in the image that contain the most contrast/content. If you have much content that is present in only one image, masking that area out will prevent it from leading the registration astray. Masking provides the opportunity to specify the important regions and make the algorithm ignore the image content outside the mask. This does not mean that the rest is not registered, but rather that it moves along passively, i.e. areas outside the mask do not actively contribute to the cost function that determines the quality of the match. Note the mask defines the areas to include, i.e. to exclude a particular region, build a mask that contains the entire object/image except that region.
- Note: masking within the registration is different from feeding a masked/stripped image as input, where areas of no interest have been erased. Such masking can still produce valuable results and is a viable option if the module in question does not provide a direct masking option. But direct masking by erasing portions of the image content can produce sharp edges that registration methods can lock onto. If the edge becomes dominant then the resulting registration will be only as good as the accuracy of the masking. That problem does not occur when using masking option within the module.
- The following modules currently (v.3.6.1) provide masking:
- General Registration (BRAINS) module : in the Mask Option tab, you can specify a mask for both fixed and moving image. Note that you need to specify both and that both need to be labelmaps with value 1 in areas you wish included and 0 elsewhere. Also both mask images must be the same size (image dimensions) as the image being registered; i.e. if you register two images of different size you cannot use the same mask for both, because the mask for the fixed image must match the fixed image size and the mask for the moving image must match the moving image size. But you can use the ResampleScalarVectorDWIVolume module to resample one mask to the resolution of the other (use the appropriate image as Reference) and then you can use it.
- Expert Automated Registration module see in the Advanced Registration Parameters tab:you can specify 1 mask for the fixed image only. Mask should be a labelmap with value 1 in areas you wish included and 0 elsewhere.
How do I generate a mask and what should it look like ?
The purpose of masking (see above) is to exclude any parts of the image that are present in only one of the images yet large enough to distract the registration algorithm as it is trying to find a match for something that doesn't exist. So your mask should focus on excluding those (irrelevant) aspects of the image. Also if you have deformations in the image and care about accuracy in one region more than another you would center the mask around that region. There is a set of screencasts dedicated to quick mask building here:
Registration Video Tutorials
Basically you can use any segmentation tool in Slicer that produces a labelmap.
Some important things to remember with masking are:
- masks need not be super accurate, since in most cases only a subset of points inside the mask is used to drive the registration.
- increase the number of sample points from what you would use for the entire image, since you will loose points that fall outside the mask
- there must be a good amount of initial overlap between the masks for the registration algorithm to find a match
- if there are prominent edges important for the registration, make sure they are inside the mask. The dilation tool in the editor is a good way to enlarge masks. The video tutorials above do show how to do that. There's a special video comparing registration with and without the dilation ("B-spline registration with masking")
- you need a mask for both the fixed and the moving image.
There are many example cases in the Slicer Registration Library that demonstrate the use of masks, e.g.:
Which registration methods offer masking?
By masking we mean to selectively include only parts of an image to
selectively participate (i.e contribute to the optimization) . Masking is supported directly by
- the General Registration (BRAINS) module
- the Expert Automated Registration )module
Note that the use of masks differs among these: "Expert Auto" requires a binary mask for fixed image only, while "General BRAINS" requires masks for both fixed and moving image. Expected is a label map with values 0 and 1, 1 being the regions to include.
Is there a function to convert a box ROI into a volume labelmap?
The Documentation/Nightly/Extensions/VolumeClip ''Volume clip with ROI box'' module can fill a volume inside a region and outside the region with two different values.
Slicer version 3.6 supports conversion of a box ROI into a labelmap via the Crop Volume module. This function has not (yet) been ported to Slicer4. You can create a new ROI box or select an existing one. You must select an image volume to crop for the operation, even if you're only interested in the ROI labelmap. You need not select a dedicated output for the labelmap, it is generated automatically when the cropped volume is produced, and will be called Subvolume_ROI_Label in the MRML tree. After creating the box ROI labelmap, simply delete the cropped volume and other output like the "...resample-scale-1.0" volume.
Likely you will need the volume with the same dimension and pixel spacing as the reference image. The box volume produced above has the correct dimension, but is only 1 voxel in size. Hence there is a second step required, which is to resample the Subvolume_ROI_Label to the same resolution: use the ResampleScalarVectorDWIVolume module and select the appropriate reference and Nearest Neighbor as interpolation method. Finally go to the Volumes module and check the Labelmap box in the info tab to turn the volume into a labelmap.
I have to manually segment a large number of slices. How can I make the process faster?
Contour every other slice, then run a Dilate and an Erode operation. For further speed increase (at the cost of losing more details), you may contour just every 3rd or 4th slice and then run Dilate multiple times (until all the holes are filled in) and then run Erode as many times as you ran Dilate. You can also subsample your image first, then edit, and finally upsample again and use the SmoothLabelmap module to reduce artifacts. See FAQ below for examples on quick segmentation methods.
How can I quickly generate a mask image ?
Several tools exist to interactively segment structures. A collection of short video examples demonstrating the methods can be found here : Video Tutorials on Mask generation
Parameters & Concept
What's the difference between Rigid and Affine registration?
Rigid registration is a transform with 6 degrees of freedom (DOF): 3 translations (one along each axis) and 3 rotations (one around each axis). An affine registration includes 12 DOF, i.e. 3 additional DOF for scaling (along each axis) and shearing. So strictly speaking an affine transform is a non-rigid transform, even though linear, because the volume can distort. However in practice by non-rigid transform one usually refers to nonlinear transforms with more than 12 DOF, e.g. BSpline or polynomial models.
What's the difference between Affine and BSpline registration?
An affine registration includes 12 DOF, i.e. 3 for translations (one along each axis), 3 rotations (one around each axis), 3 for scaling (along each axis) and 3 for shearing. So strictly speaking an affine transform is a non-rigid transform, even though linear, because the volume can distort. However in practice by non-rigid transform one usually refers to nonlinear transforms with more than 12 DOF, e.g. BSpline or polynomial models. A BSpline registration employs a nonlinear nonrigid model that allows individual regions of the image to distort independently, but enforcing smooth transitions between them. A BSpline transform is not described by a 4x4 matrix like the affine model, but by a list of displacement vectors for each point along a prescribed grid. E.g. a 3x3x3 BSpline grid has 27 points that can move independently, yielding 27 DOF; a 5x5x5 grid analogously has 125 DOF etc.
What's the difference between the various registration methods listed in Slicer?
Most of the registration modules use the same underlying ITK registration algorithm for cost function and optimization, but differ in implementation on parameter selection, initialization and the type of image toward which they have been tailored. To help choose the best one for you based on the method or available options, an.
To help choose based on a particular image type and content, you will find many example cases incl. step-by-step instructions and discussions on the particular registration challenges in the Slicer Registration Library. The library is organized in several different ways, e.g. consult this sortable table with all cases and the method used.
Note that registration methods are in continuous development, and not all cases in the Registration Library, may be updated for the latest version. To see which version they were verified on, check the icon in the title of the case page.
How many sample points should I choose for my registration?
- Problem: unsure what the Sample Points setting means or how I could use it to improve my registration.
- Explanation:All registration modules contain a parameter field that controls how much of the image is sampled when performing an automated registration. The unit is often an absolute count, but in some cases also a percentage. Default settings also vary among modules. The number of samples is an important setting that determines both registration speed and quality. If the sample number is too small, registration may fail because it is driven by image content that insufficiently represents the image. If sample number is too large, registration can slow down significantly.
- Fix: If registration speed is not a major issue, better to err on the side of larger samples. Most default settings are chosen to yield relatively fast registrations and for most of today's image represent only a small percentage. Below the defaults for the different registration modules, for version 3.6.1:
- Defaults Used in Slicer Modules v.3.6.1
- BSpline Deformable Registration module: 50,000
- Expert Automated module Rigid 1%; Affine 2%, BSpline 10%
- General Registration (BRAINS) module: 100,000
The table below relates total sample points and percentages to the most common image sizes. Also consider that sample points are chosen randomly so that some points may fall outside the actual object to be registered. That is not a bad thing per se, some background points are important, but not if they are too far from the edges of the object. So consider both total image size as well as the percentage of the image field of view that your object of interest obtains. E.g. if your object fills only half the image, double the sample points to get the desired amount of points within the object.
| Image Size | Total Points | 10,000 | 50,000 | 100,000 | 200,000 | 1% | 2% | 5% | 10% | 20% |
|---|---|---|---|---|---|---|---|---|---|---|
| 128 x 128 x 64 | 1048576 | 1.0% | 4.8% | 9.5% | 19.1% | ~10000 | ~20000 | ~52500 | ~100000 | ~200000 |
| 256 x 256 x 30 | 1966080 | 0.5% | 2.5% | 5.1% | 10.2% | ~20000 | ~40000 | ~97500 | ~200000 | ~400000 |
| 256 x 256 x 120 | 7864320 | 0.1% | 0.6% | 1.3% | 2.5% | ~77500 | ~150000 | ~400000 | ~775000 | ~1500000 |
| 192 x 192 x 192 | 7077888 | 0.1% | 0.7% | 1.4% | 2.8% | ~70000 | ~150000 | ~350000 | ~700000 | ~1500000 |
| 512 x 512 x 48 | 12582912 | 0.1% | 0.4% | 0.8% | 1.6% | ~125000 | ~250000 | ~625000 | ~1250000 | ~2500000 |
| 256 x 256 x 256 | 16777216 | 0.1% | 0.3% | 0.6% | 1.2% | ~175000 | ~325000 | ~850000 | ~1750000 | ~3250000 |
| 512 x 512 x 160 | 41943040 | 0.0% | 0.1% | 0.2% | 0.5% | ~425000 | ~850000 | ~2000000 | ~4250000 | ~8500000 |
The image below shows sample point densities on one slice of a brain MRI with 256x256x130 voxels. For robust registration, we recommend a useful coverage requires at least 1% coverage for affine, more (at least 5%) for nonrigid (BSpline) as DOF increase:
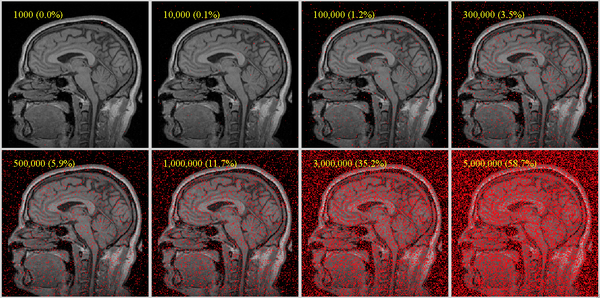
Can I register 2D images with Slicer?
Slicer is designed to work with 3D images. Since it operates in physical space, all objects must have a physical dimension (e.g. thickness).
It is possible to still run a 2D registration with the 3D tools, with limitations. The two options are:
1.manual registration: the manual registration can easily be restricted to 2D. Simply do not use the sliders that control out-of plane rotation or translation. For more info on manual registration see FAQ on manual registration and also the Registration Library
2.Automated registration: the basic concept here is to pad the image with duplicated slices in the 3rd dimension to provide the appearance of a 3D object (e.g. using the Resample module), and then try to lock all degrees of freedom pertaining to the 3rd dimension (e.g. by masking), i.e. out-of-plane rotation or translation. Because the amount of data is very limited in 2D, automated registration is likely going to fail if the images are too far apart or to dissimilar in content.
An example of 2D registration can be found in our case library:
https://www.slicer.org/wiki/Documentation:Nightly:Registration:RegistrationLibrary:RegLib_C46 === What's the BSpline Grid Size? ===
The BSpline grid is the basic structure of the deformation model applied when transforming one image to match the other. A uniform grid is placed over the entire image and the grid-points are allowed to move freely. These node displacements are then interpolated over the remainder of the grid. The image below shows an example of a 3x3x3 grid. An additional row of grid points is created at the boundary of the image (red). Those points are locked and do not move. They provide a boundary constraint to keep the grid from shifting. The interior points (green) are free to move in any direction (blue arrows), which gives 3 degrees of freedom (DOF) per grid-point. So a 3x3x3 grid translates into a transform with 27 DOF, a 5x5x5 grid will have 125 DOF. For comparison a rigid transform has 6 DOF, an affine transform has 12.
When specifying a grid, be aware of your image resolution and anisotropy. For example if your image has substantially fewer voxels in one direction, it may be wise to also request fewer grid-points in that dimension, to avoid over fitting.
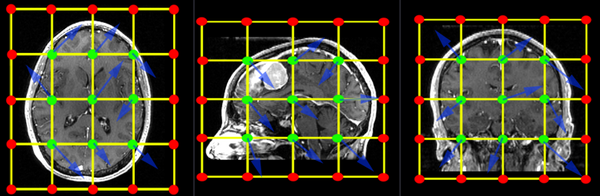
I want to register two images with different intensity/contrast.
The two most critical image features that determine automated registration accuracy and robustness are image contrast and resolution. Differences in image contrast are best addressed with the appropriate cost function. The cost function that has proven most reliable for registering images with different contrast (e.g. a T1 MRI to a T2 or an MRI to CT or PET) is mutual information. All intensity-based registration modules use mutual information as the default cost function. Only the Expert Automated Registration module (under All Modules) lets you choose alternative cost function. No extra adjustment is therefore needed in terms of adjusting parameters to register images of different contrast. Depending on the amount of differences you may consider masking to exclude distracting image content or adjusting the Histogram Bins setting to increase/decrease the level of intensity detail the algorithm is aware of.
- Modules that allow control of Histogram Bins:
How important is bias field correction / intensity inhomogeneity correction?
While registration may still succeed with mild cases of inhomogeneity, moderate to severe inhomogeneity can negatively affect automated registration quality. It is recommended to run an automated bias-field correction on both images before registration.This becomes more relevant the more degrees of freedom you have in your registration, i.e. a BSpline registration is more likely to be affected by an intensity bias than an Affine. The image here (click to enlarge)  shows an example of a low-level BSpline deformation applied once with and once without preceeding bias-correction. You can see how the lack of signal at the bottom of the uncorrected image causes the image to be "pulled downward:m which is incorrect.
see here for documentation on the BiasCorrection module. Masking of non-essential peripheral structures can also help to reduce distracting image content. See the FAQ on masking for more details.
shows an example of a low-level BSpline deformation applied once with and once without preceeding bias-correction. You can see how the lack of signal at the bottom of the uncorrected image causes the image to be "pulled downward:m which is incorrect.
see here for documentation on the BiasCorrection module. Masking of non-essential peripheral structures can also help to reduce distracting image content. See the FAQ on masking for more details.
Have the Slicer registration methods been validated?
The Slicer3.6 registration modules share the same same underlying ITK registration engines. For validation of those basic algorithms refer to the ITK software guide or the Insight Journal. Slicer registration of images and surfaces has been applied successfully in many cases, some of which are documented here here. For ongoing efforts on improving/validating Slicer registration performance please contact the Slicer User Mailing List or the Slicer Developer Mailing List.
How can I save the parameter settings I have selected for later use or sharing?
The Parameter Set menu at the top tab of each module serves this purpose of saving and recalling instances of parameter settings. To save the current settings select Rename from the menu, give it a descriptive name and then select Create New Commandline Module from the menu. New settings are added to the menu as created. These settings are saved together with the Slicer Scene.mrml file. To save only the presets without any associate data, first save your entire scene, then delete all nodes in the MRML tree except the top Scene node, and then save this under a new name like "Slicer_Presets.mrml" etc. To load the presets in, use Import from the File menu, which will add the MRML entries to the existing scene. Do not load presets via "Load Scene", since that will replace the loaded data rather than add to it. A detailed user guide on loading presets can be found here. Note that the content and format of the Slicer Scene file tends to change and evolve with newer versions, and presets may not be compatible across very different versions.
Registration is too slow. How can I speed up my registration?
The key parameters that influence registration speed are the number of sample points, the degrees of freedom of the transform, the type of similarity metric and the initial misalignment and image contrast/content differences. If registration quality is ok, try reducing the sample points first. Guidelines on selecting sample points are given here. The degrees of freedom usually are given by the overall task and not subject to variation, but depending on initial misalignment, both speed and robustness can improve by an iterative approach that gradually increases DOF rather than starting with a high DOF setting. The General Registration (BRAINS) module and Expert Automated Registration module both allow prescriptions of iterative DOF.
If using a cost/criterion function other than mutual information, note that the corresponding ITK implementation may not have been parallelized and hence may not be taking advantage of multi-threading/multi-CPU cores on your computer. This can slow down performance significantly..
General tips:
- the General Registration (BRAINS) module is parallelized and tends to be fastest
- try reducing the number of sample points; see here for guidelines on selecting sample points
- try a lower DOF registration first to determine if initial position differences are causing excessive search that slows down registration.
- use mutual information as cost function (default)
- if your image is very large (512x512 in plane or greater) try a lower resolution version of your image. You can use the ResampleScalarVolume module to subsample your image.
How do I register images via landmarks/fiducials?
There are two fiducial registration tools available within Slicer. The default is within the Registration menu, and an alternative version is found in the SlicerIGT extension as "Fiducial Registration Wizard". A detailed case example using both tools is available in the Case 48 in the Registration Case Library. A brief screencast on selecting landmarks and running either tools are also here:
- fiducial-based registration
- alternative tool: using SlicerIGT's fiducial-based registration (shows extension download and module execution only, for the landmark selection see movie above)
Is the BRAINSfit registration for brain images only?
No, it is applicable and has been used successfully on non-brain image data. See the Registration Case Library for examples. The BRAINS name is derived from the ICTS at program at the University of Iowa, where it was developed, details of which you will find here.
One of my images has a clipped field of view. Can I still use automated registration?
Probably yes. Best remedy is to apply a mask that excludes the missing portion in the other image. Also initialization is critical, more so than with full FOV images. If images have large amount of initial misalignment, try to center both first (see here for details) or perform a cursory manual alignment (see here for details) and use that as initialization. An example of registrations with clipped FOV can be found in the Slicer Registration Library , e.g. case 4, case9; case 13; case 30 and others.
Can I combine multiple registrations?
Yes, you can nest multiple (affine) registrations inside eachother. You can generate combined ones via the right-click context menu in the Data module and selecting Harden Transform. Note that the original transform or volume is replaced when selecting Harden Transform, so it is recommended to rename the node afterwards to document the fact.
Currently (v.4.1) BSpline transforms cannot be combined with other transforms, so if you have combinations of Affine and BSpline or multiple BSpline transforms you need to resample multiple times to apply them all. We recommend to supersample the volume beforehand to counteract interpolation blurring.
Is there a module for surface registration?
The current (4.5) Slicer core does not include a surface registration, but available in extensions.
- Rigid, similarity or affine registration of model to model: SlicerIGT extension, Model registration module (in IGT category)
- Rigid, similarity or affine registration of markups fiducials to model: SlicerIGT extension, Fiducials-Model registration module (in IGT category)
Can I combine image and surface registration?
Not simultaneously. You can run surface and image registrations separately and then combine the transforms, but there is currently no support to combine both surface and intensity data into a single cost function.
Apply, Resample, Export
I ran a registration but cannot see the result. How do I visualize the result transform?
There are 2 ways to see the result of a registration: 1) by creating a new resampled volume that represents the moving image in the new orientation , 2) by direct (dynamic) rendering of the original image in a new space when placed inside a transform. The latter is not available for non-rigid transforms, hence a registration that includes nonlinear components (BSpline, Warp) must be visualized by first resampling the entire volume with the new transform. If you ran a registration yet see no effect, the reason could be one of the following:
- you did not request an output. In the module parameters section, you must specify either an output transform or an output image/volume. Select one or both (for nonrigid registration).
- you requested a result transform but the moving volume is not placed inside the transform in the MRML tree. Not all modules automatically place the moving image inside the result transform node. To see the effect do the following:
- Go to the Data module
- left click on the volume you wish to register, and drag & drop it onto the new transform node created by your registration. After dropping it once the transform, you should see a "+" appear in front of the transform node, and clicking on it should reveal the volume now inside/under that transform.
- make sure you have the image for which you wish to adjust the registration selected and visible in the slice views, preferably all 3 views (sagittal, coronal, axial). If the image is visible in the Slicer views, you should see a change immediately when the volume is placed inside the transform
- registration completed with an error. Check for status messages at the top of the module and look at the Error Log in the window menu to see if there were any errors reported.
- registration completed with a result that is too small to be noticeable: For linear transforms (rigid to affine) you can look at and edit the transform directly in Slicer: go to the Transforms module and select the transform. You should see the parameters in the 4x4 matrix displayed. If diagonal numbers are very close to 1 and all others very close to 0, you have a transform that barely moves the object at all. Nonrigid transforms you have to save to a file first; you can then view the .tfm file with any text editor.
- you performed a non-rigid (BSpline) transform but did not yet request an output volume.
My reoriented image returns to original position when saved; Problem with the Harden Transform function
You can apply an affine transform to an image by creating a transform, placing the volume inside that transform in the Data module, and then selecting Harden Transform via the context-menu (right click on the image volume). This will move the image back out to the main level and "apply" the transform. It will, however, not resample the image data, but rather place the information about the new orientation into the image header. When the image is saved, this information is saved also as part of the file header, as long as orientation data is supported by the file format. If the saved volume is now loaded by another software that does not consider this header orientation (e.g. ImageJ) or does not visualize the image in physical space, then the image will appear in its old position.
While resampling of data is best avoided if possible, a hard resampling is possible through the ResampleScalarVectorDWIVolume module: select your image and transform as input, create a new volume as output and click Apply. This new volume will now be in the new orientation that will be retained if saved and reloaded elsewhere.
Resampling with the Resample Image BRAINS module:
To get your moving image in the same orientation AND the same pixel matrix as your fixed/target image, you need to resample it with the fixed image as reference:
- go to the Registration / Resample Image (BRAINS) module
- for "Image To Warp" , select your moving image
- for "Reference Image" select your fixed image (i.e. the image whose resolution you wish to match)
- for "Output Image" select" create new & rename
- for "Transform file": select the manual registration transform file you created
What is the Meaning of 'Fixed Parameters' in the transform file (.tfm) of a BSpline registration ?
A typical BSpline transform file will contain 2 transforms, an affine portion (commonly saved as "Transform 1" at the end of the file), and a nonrigid BSpline portion (commonly saved as "Transform 0"). The bulk of the BSpline part are 3D displacement vectors for each of the BSpline grid-nodes in physical space, i.e. for each grid-node, there will be three blocks of displacements defining dx,dy,dz for all grid nodes. After this field is a "Fixed Parameters" section that may look like this:
FixedParameters: 8 8 8 -54.1406 -54.1406 -35 54.1406 54.1406 35 1 0 0 0 1 0 0 0 1
The first 3 numbers are the actual grid size (number of knots in each dimension), which is always larger than your requested grid because the grid is extended beyond the image margin to prevent clipping. The next 3 numbers is the origin of the grid, spacing of the grid, and the direction cosines of the grid. More details on the format in the ITK documentation (ITKSoftwareGuide.pdf)
After registration the registered image appears cropped. How can I increase the field of view to see/include the entire image
Problem: The registration results seems to be correct but the output volume shows only the overlapping region. Is there a way that to show the whole registered volume including the parts which are not overlapping? There's two answers to this problem: one is a visualization issue related to how Slicer chooses the field of view (FOV), and one is how the FOV is chosen for resampling. Slice views: Slicer sets the overall field of view based on the image selected as "background". If the image in the foreground extends beyond that region it will be clipped. You can easily fix/test that by switching foreground and background volumes. If you already created a resampled version of the registered image, that may also have been cropped,because it sets the field of view based on the reference image (which is the fixed image usually). If you place your resampled image in the background and still see cropped edges, then that's what happened. In that case either use a different reference image for resampling or no reference at all and specify the result sizes manually, or use the crop tool to expand the FOV as described below:
- use the CropVolume module to over-crop the reference image (define ROI to cover the field of view you want to have in the resampled volume). You will need to use interpolated crop mode, so the resolution will not match the original reference volume.
- Use the result of the cropping operation as the reference for BRAINSResample module (you will need to specify the moving volume, output volume, and the transform produced by the BRAINS registration module).
How can I see the parameters of the function that describe a BSpline registration/deformation?
To see the parameters of the transform, you have to write it to file and investigate by other means. The BSpline transform is saved as a ITK .tfm which is a text file containing the displacement vectors of each grid-point, plus any initial affine transform (if present). One nice way to visualize is to create a grid image of the same dimensions as your target, and then apply the transform to this grid image, you can then see the deformations as deformations in the gridlines. Example Grid images can be downloaded here. Use the ResampleScalarVectorDWIVolume module for the resampling.
A quick way directly in slicer is to place the undeformed and deformed volumes into back- and foreground and fade back and forth with the fading slider. Another alternative is to convert the transform into a 4-D deformation field directly and visualize it in slicer using RGB color. See FAQ below on how to convert.
How to export the displacement magnitude of the transform as a volume?
This is possible but requires use of the interactive python console:
transformNode=slicer.util.getNode('LinearTransform_3')
referenceVolumeNode=slicer.util.getNode('MRHead')
slicer.modules.transforms.logic().CreateDisplacementVolumeFromTransform(transformNode, referenceVolumeNode, False)
The new volume can then be saved or used for ROI analysis with the Label Statistics module.
Physical Space vs. Image Space: how do I align two registered images to the same image grid?
Slicer displays all data in a physical coordinate system. Hence an image can only be displayed correctly if it contains sufficient header information to relate the image voxel grid with physical space. This includes voxel size, axis orientation and scan order. It is therefore possible for two images to be aligned when viewed in Slicer, even though their underlying image grid is oriented very differently. To match the two images in image as well as physical space, the abovementioned axis direction, voxel size and image grid orientation must match. The procedure will depend on the image data, but the main tools at your disposal are the ResampleScalarVectorDWIVolume module and OrientImages module.
Nonrigid / BSpline Registration
The nonrigid (BSpline) registration transform does not seem to be nonrigid or does not show up correctly.
See FAQ on viewing registration results here. BSpline transforms are not available for immediate rendering by placing volumes or models inside the transforms. Only linear transforms can be viewed that way. A BSpline transform must be visualized by resampling the entire volume with the new transform. If you did not yet an output volume when running the registration, you can do so after the fact, use the ResampleScalarVectorDWIVolume module.
What's the difference between BRAINSfit and BRAINSDemonWarp?
General Registration (BRAINS) module performs Rigid to Affine and BSpline registration that commonly will have at the most a few hundred degrees of freedom (DOF), whereas the Demon Registration (BRAINS) module performs a optic flow high-DOF warping scheme that has many thousands of DOF and is significantly less constrained.
How can I convert a BSpline transform into a deformation field?
There is a dedicated module to convert a BSpline ITK transform file (.tfm) into a deformation field volume.
Modules -> all Modules -> BSpline to deformation field
provide your BSpline transform and a reference image that defines the size of the deformation field.
For Slicer 3.6 this functionality was available as command line functionality only:
To execute, type (exchange /Applications/Slicer3.6.3 with the path of your Slicer installation):
/Applications/Slicer3.6.3/Slicer3 --launch /Applications/Slicer3.6.3/lib/Slicer3/Plugins/BSplineToDeformationField --tfm InputBSpline.tfm --refImage ReferenceImage.nrrd --defImage Output_DeformationField.nrrd
for more details try:
/Applications/Slicer3.6.3/Slicer3 --launch /Applications/Slicer3.6.3/lib/Slicer3/Plugins/BSplineToDeformationField --help
Where can I check the reference color map for the transform visualizer? i.e. how can I know the magnitude of transform vectors?
The "Transform Visualizer" is a tool that in earlier Slicer versions (4.2 and before) was an extension module, but in newer versions has been integrated into the Transforms module] (under the Display tab). It enables visualisation of transform vectors that are particularly useful to help understand complex non-rigid deformations such as B-Spline registrations.
You can find the referenced color table in the Colors module . There is a category called “Transform Display” and under that there is the “Displacement to color” color table. To display the scalar value represented by a color, you need to turn on the “Display scalar bar” option in Scalar Bar section under “Colors” module. Currently it only shows the scalar bar in the 3D window.
Alternatlively, you can see the deformation vector under current cursor in Transform module in the Information section, when you move your cursor, it will display the current vector.