Difference between revisions of "Documentation/Nightly/Extensions/MABMIS"
XiaofengLiu (talk | contribs) (Created page with '<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- -----------------------…') |
XiaofengLiu (talk | contribs) |
||
| (15 intermediate revisions by the same user not shown) | |||
| Line 8: | Line 8: | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
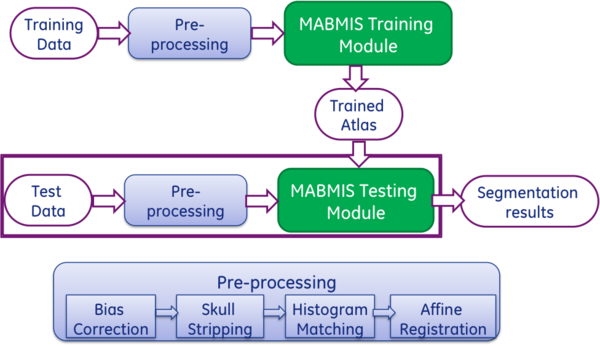
| + | MABMIS is a Slicer extension that implements a multi-atlas based multi-image method for group-wise segmentation [1]. The method utilizes a novel tree-based groupwise registration method for concurrent alignment of both the atlases and the target images, and an iterative groupwise segmentation method for simultaneous consideration of segmentation information propagated from all available images, including the atlases and other newly segmented target images. | ||
| + | <br> | ||
Extension: [[Documentation/{{documentation/version}}/Extensions/MABMIS|MABMIS: Multi-atlas based group-wise segmentation]]<br> | Extension: [[Documentation/{{documentation/version}}/Extensions/MABMIS|MABMIS: Multi-atlas based group-wise segmentation]]<br> | ||
| − | Acknowledgments: This project was supported by NIH grant | + | Acknowledgments: This project was supported by NIH grant 2R01EB006733-04A1. <br> |
Contributor: Xiaofeng Liu, Minjeong Kim, Jim Miller, Dinggang Shen. <br> | Contributor: Xiaofeng Liu, Minjeong Kim, Jim Miller, Dinggang Shen. <br> | ||
Contact: Xiaofeng Liu, <email>xiaofeng.liu@ge.com</email><br> | Contact: Xiaofeng Liu, <email>xiaofeng.liu@ge.com</email><br> | ||
| − | {{documentation/{{documentation/version}}/ | + | {{documentation/{{documentation/version}}/module-introduction-row}} |
| + | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| + | |{{collaborator|logo|unc}}|{{collaborator|longname|unc}} | ||
| + | |{{collaborator|logo|ge}}|{{collaborator|longname|ge}} | ||
| + | }} | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| + | <!-- ---------------------------- --> | ||
| + | |||
| + | |||
| + | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| + | <!-- ---------------------------- --> | ||
| + | {| | ||
| + | |[[Image:MABMIS_algorithm.png|thumb|600px|MABMIS algorithm overview. ]] | ||
| + | |} | ||
| + | |||
| + | The figure above shows an overview of the algorithm. MABMIS contains two modules: the training module, and the testing module. | ||
| + | <br> | ||
| + | |||
| + | In the training module, multiple atlas image pairs are required as the input. Each atlas image pair contains one pre-processed T1-weighted intensity image and one label image on which each interested structure on the T1 image is manually identified and assigned a unique label. The label image is generally generated by an expert. In the training module, the multiple atlas pairs are processed to construct an atlas tree, where each atlas is one node of the tree. The tree is constructed based on the similarity among the atlases and tree-based groupwise registration. | ||
| + | <br> | ||
| + | |||
| + | In the testing module, a group of target images are segmented based on the atlas tree. First, a novel tree-based groupwise registration method is employed to simultaneously register them to the atlas tree, the target images are segmented simultaneously using an iterative groupwise segmentation strategy, which provides improved accuracy and across-image consistency. | ||
| + | <br> | ||
| + | |||
| + | Before applying the modules, both atlas images and target images need to be pre-processed. The processing steps include bias correction ( e.g., N4), skull stripping (e.g., skullstripper in slicer), histogram matching. All images are then registered to a common space using affine registration. The pre-processing tools are not included in the module. | ||
| + | |||
| + | <br> | ||
| + | Detailed description of the algorithm can be found in [1]. | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
| + | {| | ||
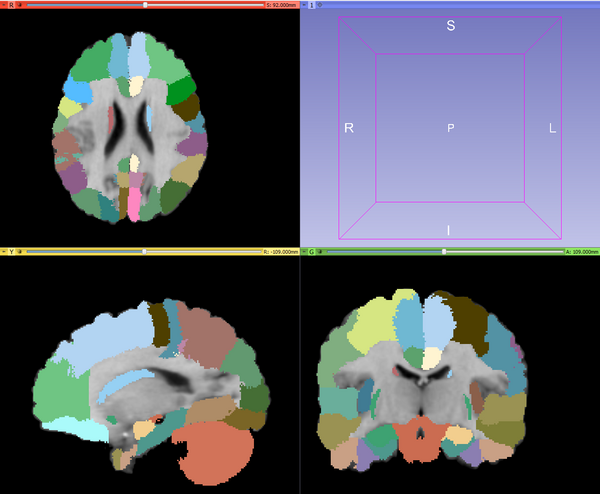
| + | |[[Image:MABMIS_results.png|thumb|600px|Example: MABMIS segmentation results]] | ||
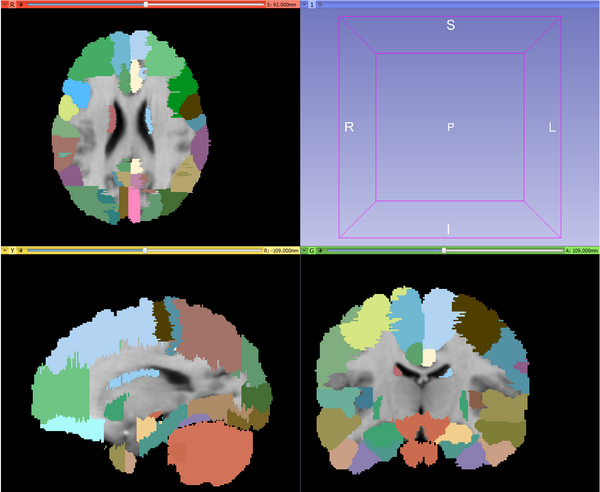
| + | |[[Image:MABMIS_groundtruth.png|thumb|600px|Example: The ground truth]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| + | The module has two parts: training and testing. The training module requires as input a set of atlases, where each atlas contains an intensity image and a pre-segmented label images. The module then generates a trained atlas tree that can be used for multi-structure segmentation in the testing phase. The testing module requires two inputs. The first one is the trained atlas tree, and the second one is the set of images to be segmented. As a result, the testing module outputs multi-structure segmentation results on these images. | ||
| + | <br> | ||
| + | |||
| + | == Training == | ||
| + | |||
| + | The GUI for the training module is shown below: | ||
| + | {| | ||
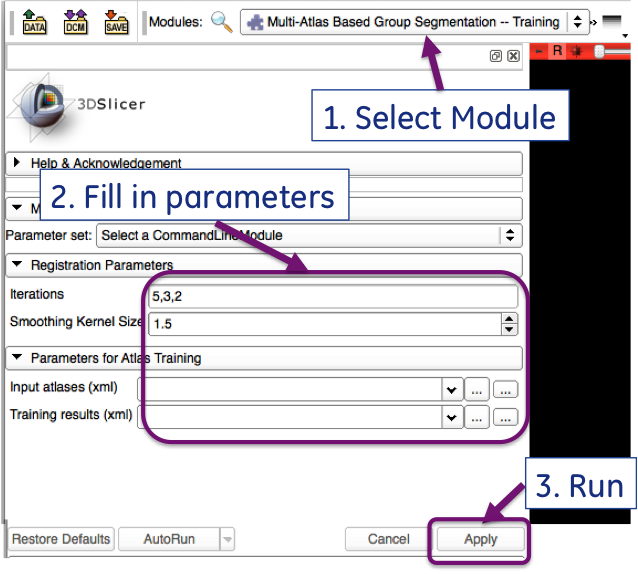
| + | |[[Image:MABMIS_trainning_GUI.png|thumb|800px|MABMAS training module]] | ||
| + | |} | ||
| + | |||
| + | <br> | ||
| + | |||
| + | To run it as a command line: | ||
| + | <br> | ||
| + | |||
| + | '''IGR3D_MABMIS_Training''' ''[-i xx,xx,xx] [-s sigma] -- trainingXML TrainingData.xml -- atlasTreeXML TrainedAtlas.xml '' | ||
| + | <br> | ||
| + | |||
| + | * [-i xx,xx,xx]:(OPTIONAL) number of iterations used in image registration. It is three numbers separated by comma. Default value is 5,3,2. | ||
| + | * [-s sigma]: (OPTIONAL)the size of the smoothing kernel that is used for smoothing deformation fields. Default value is 1.5. | ||
| + | * TrainingData.xml : an xml file that specifies information about the training images, including the number of training data, filenames of T1-weighted image and the segmented label images. An example can be found in the test folder. | ||
| + | * TrainedAtlas.xml: an xml file for output, that stores information about the trained atlas tree which can be used to segment. | ||
| + | |||
| + | == Testing == | ||
| + | |||
| + | The GUI for the testing module is shown below: | ||
| + | {| | ||
| + | |[[Image:MABMIS_testing_GUI.png|thumb|600px|MABMAS testing module]] | ||
| + | |} | ||
| + | |||
| + | <br> | ||
| + | |||
| + | To run it as a command line: | ||
| + | <br> | ||
| + | |||
| + | '''IGR3D_MABMIS_Testing''' ''[-i xx,xx,xx] [-s sigma] --atlasTreeXML TrainedAtlas.xml -- imageListXML TestImageList.xml --outputfolder OutputFolder'' | ||
| + | |||
| + | <br> | ||
| + | |||
| + | * [-i xx,xx,xx]: (OPTIONAL) number of iterations used in image registration. It is three numbers separated by comma. Default value is 5,3,2. | ||
| + | * [-s sigma]: (OPTIONAL)the size of the smoothing kernel that is used for smoothing deformation fields. Default value is 1.5. | ||
| + | * TrainedAtlas.xml: an xml file that contains the trained atlas resulted from the training module. | ||
| + | * TestImageList.xml: an xml file that contains a list of test images to be segmented. An example can be found in the test folder | ||
| + | * OutputFolder: The location to save the segmentation results. | ||
| + | |||
| + | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
| + | |||
| + | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| + | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|References}} | ||
| + | *Hongjun Jia, Pew-Thian Yap, Dinggang Shen, "Iterative multi-atlas-based multi-image segmentation with tree-based registration", NeuroImage, 59:422-430, 2012. | ||
| + | |||
| + | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
| + | {{documentation/{{documentation/version}}/module-developerinfo}} | ||
| + | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-footer}} | ||
| + | [[Category:Documentation/{{documentation/version}}/Modules/Segmentation]] | ||
| + | <!-- ---------------------------- --> | ||
Latest revision as of 14:29, 17 January 2014
Home < Documentation < Nightly < Extensions < MABMIS
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
MABMIS is a Slicer extension that implements a multi-atlas based multi-image method for group-wise segmentation [1]. The method utilizes a novel tree-based groupwise registration method for concurrent alignment of both the atlases and the target images, and an iterative groupwise segmentation method for simultaneous consideration of segmentation information propagated from all available images, including the atlases and other newly segmented target images.
Contributor: Xiaofeng Liu, Minjeong Kim, Jim Miller, Dinggang Shen. | |||||
|
Module Description
The figure above shows an overview of the algorithm. MABMIS contains two modules: the training module, and the testing module.
In the training module, multiple atlas image pairs are required as the input. Each atlas image pair contains one pre-processed T1-weighted intensity image and one label image on which each interested structure on the T1 image is manually identified and assigned a unique label. The label image is generally generated by an expert. In the training module, the multiple atlas pairs are processed to construct an atlas tree, where each atlas is one node of the tree. The tree is constructed based on the similarity among the atlases and tree-based groupwise registration.
In the testing module, a group of target images are segmented based on the atlas tree. First, a novel tree-based groupwise registration method is employed to simultaneously register them to the atlas tree, the target images are segmented simultaneously using an iterative groupwise segmentation strategy, which provides improved accuracy and across-image consistency.
Before applying the modules, both atlas images and target images need to be pre-processed. The processing steps include bias correction ( e.g., N4), skull stripping (e.g., skullstripper in slicer), histogram matching. All images are then registered to a common space using affine registration. The pre-processing tools are not included in the module.
Detailed description of the algorithm can be found in [1].
Use Cases
Tutorials
The module has two parts: training and testing. The training module requires as input a set of atlases, where each atlas contains an intensity image and a pre-segmented label images. The module then generates a trained atlas tree that can be used for multi-structure segmentation in the testing phase. The testing module requires two inputs. The first one is the trained atlas tree, and the second one is the set of images to be segmented. As a result, the testing module outputs multi-structure segmentation results on these images.
Training
The GUI for the training module is shown below:
To run it as a command line:
IGR3D_MABMIS_Training [-i xx,xx,xx] [-s sigma] -- trainingXML TrainingData.xml -- atlasTreeXML TrainedAtlas.xml
- [-i xx,xx,xx]:(OPTIONAL) number of iterations used in image registration. It is three numbers separated by comma. Default value is 5,3,2.
- [-s sigma]: (OPTIONAL)the size of the smoothing kernel that is used for smoothing deformation fields. Default value is 1.5.
- TrainingData.xml : an xml file that specifies information about the training images, including the number of training data, filenames of T1-weighted image and the segmented label images. An example can be found in the test folder.
- TrainedAtlas.xml: an xml file for output, that stores information about the trained atlas tree which can be used to segment.
Testing
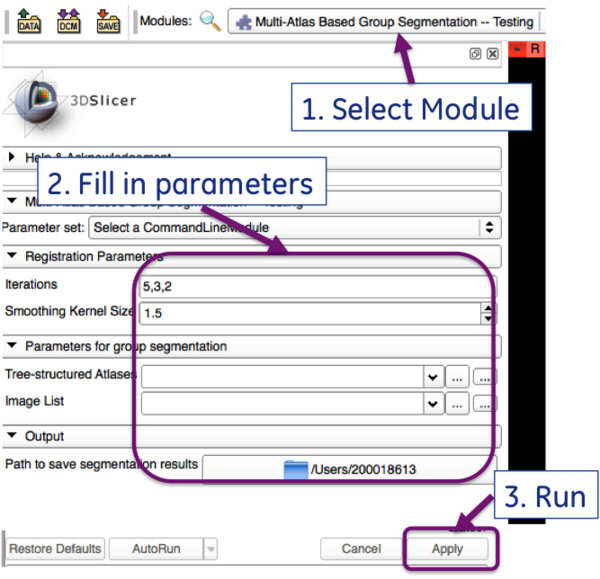
The GUI for the testing module is shown below:
To run it as a command line:
IGR3D_MABMIS_Testing [-i xx,xx,xx] [-s sigma] --atlasTreeXML TrainedAtlas.xml -- imageListXML TestImageList.xml --outputfolder OutputFolder
- [-i xx,xx,xx]: (OPTIONAL) number of iterations used in image registration. It is three numbers separated by comma. Default value is 5,3,2.
- [-s sigma]: (OPTIONAL)the size of the smoothing kernel that is used for smoothing deformation fields. Default value is 1.5.
- TrainedAtlas.xml: an xml file that contains the trained atlas resulted from the training module.
- TestImageList.xml: an xml file that contains a list of test images to be segmented. An example can be found in the test folder
- OutputFolder: The location to save the segmentation results.
Panels and their use
Similar Modules
References
- Hongjun Jia, Pew-Thian Yap, Dinggang Shen, "Iterative multi-atlas-based multi-image segmentation with tree-based registration", NeuroImage, 59:422-430, 2012.
Information for Developers
| Section under construction. |