Difference between revisions of "Documentation/4.3/Modules/PkModeling"
(Nightly -> 4.3) |
|||
| Line 10: | Line 10: | ||
Extension: [[Documentation/{{documentation/version}}/Extensions/PkModeling|PkModeling]]<br> | Extension: [[Documentation/{{documentation/version}}/Extensions/PkModeling|PkModeling]]<br> | ||
Acknowledgments: | Acknowledgments: | ||
| − | This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research.<br> | + | This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, and by National Cancer Institute as part of the Quantitative Imaging Network initiative (U01CA151261).<br> |
Implementation of the pharmacokinetics modeling was contributed by Yingxuan Zhu and Jim Miller from GE Research.<br> | Implementation of the pharmacokinetics modeling was contributed by Yingxuan Zhu and Jim Miller from GE Research.<br> | ||
Author: Yingxuan Zhu (while at GE), Andrey Fedorov (SPL), Jim Miller ({{collaborator|name|ge}})<br> | Author: Yingxuan Zhu (while at GE), Andrey Fedorov (SPL), Jim Miller ({{collaborator|name|ge}})<br> | ||
Latest revision as of 23:46, 28 October 2013
Home < Documentation < 4.3 < Modules < PkModeling
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: PkModeling | |||||
|
Module Description
|
PkModeling (Pharmacokinetics Modeling) calculates quantitative parameters from Dynamic Contrast Enhanced DCE-MRI images. This module performs two operations:
|
Use Cases
Tutorials
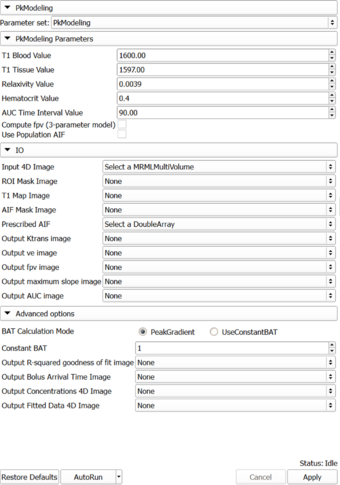
Panels and their use
|
The following acquisition parameters should be available in the NRRD header of the input data (if you are analyzing a DICOM time series, they will typically be extracted from the DICOM data):
- TR Value: Repetition time (milliseconds)
- TE Value: Echo time (milliseconds)
- FA Value: Flip angle (degrees)
- Timestamps for the dynamic series (in milliseconds)
Here is an example how this information is represented in the NRRD header:
MultiVolume.DICOM.EchoTime:=2.93 MultiVolume.DICOM.FlipAngle:=10 MultiVolume.DICOM.RepetitionTime:=6.13 MultiVolume.FrameIdentifyingDICOMTagName:=AcquisitionTime MultiVolume.FrameIdentifyingDICOMTagUnits:=ms
Similar Modules
References
- [1] Knopp MV, Giesel FL, Marcos H et al: Dynamic contrast-enhanced magnetic resonance imaging in oncology. Top Magn Reson Imaging, 2001; 12:301-308.
- [2] Rijpkema M, Kaanders JHAM, Joosten FBM et al: Method for quantitative mapping of dynamic MRI contrast agent uptake in human tumors. J Magn Reson Imaging 2001; 14:457-463.
- [3] de Bazelaire, C.M., et al., MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3.0 T: preliminary results. Radiology, 2004. 230(3): p. 652-9.
- [4] Pintaske J, Martirosian P, Graf H, Erb G, Lodemann K-P, Claussen CD, Schick F. Relaxivity of Gadopentetate Dimeglumine (Magnevist), Gadobutrol (Gadovist), and Gadobenate Dimeglumine (MultiHance) in human blood plasma at 0.2, 1.5, and 3 Tesla. Investigative radiology. 2006 March;41(3):213–21.
Information for Developers
| Section under construction. |