Difference between revisions of "Documentation/Nightly/Extensions/SlicerRT"
(Changed layout) |
m (Text replacement - "\[http:\/\/www\.slicer\.org\/slicerWiki\/index\.php\/([^ ]+) ([^]]+)]" to "$2") |
||
| (46 intermediate revisions by 7 users not shown) | |||
| Line 3: | Line 3: | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| − | [[ | + | [[{{collaborator|logo|cco}}]] |
| − | [[ | + | [[{{collaborator|logo|canarie}}]] |
| − | [[ | + | [[{{collaborator|logo|ocairo}}]] |
| + | [[{{collaborator|logo|sparkit}}]] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | {{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | ||
| − | Authors: <b>Csaba Pinter</b> (PerkLab, Queen's University), <b>Andras Lasso</b> (PerkLab, Queen's University) | + | Authors: <b>Csaba Pinter</b> (PerkLab, Queen's University), <b>Andras Lasso</b> (PerkLab, Queen's University)<br> |
| − | + | Contributors: <b>Greg Sharp</b> (Massachusetts General Hospital), Kevin Wang (Princess Margaret Hospital, UHN Toronto), Steve Pieper (Isomics)<br> | |
Contacts: | Contacts: | ||
| + | * Csaba Pinter, <email>pinter.csaba@gmail.com</email> | ||
* Andras Lasso, <email>lasso@cs.queensu.ca</email> | * Andras Lasso, <email>lasso@cs.queensu.ca</email> | ||
| − | * | + | * [https://discourse.slicer.org Slicer Forum] |
| − | * [ | + | * [[Documentation/SlicerRT/HowToReportAnError|How to report an error]] |
Website: [http://slicerrt.github.io slicerrt.org]<br> | Website: [http://slicerrt.github.io slicerrt.org]<br> | ||
License: [http://www.slicer.org/pages/LicenseText Slicer license]<br> | License: [http://www.slicer.org/pages/LicenseText Slicer license]<br> | ||
| − | '''Download/install:''' install 3D Slicer, start 3D Slicer, open the Extension Manager, install the SlicerRT extension (see more details [ | + | '''Download/install:''' install 3D Slicer, start 3D Slicer, open the Extension Manager, install the SlicerRT extension (see more details [http://slicerrt.github.io/Download.html on the download page]) |
<!-- | <!-- | ||
| Line 29: | Line 31: | ||
{| | {| | ||
| | | | ||
| − | [[Image: | + | [[Image:SlicerRT_Logo_3.0_128x128.png]] |
| | | | ||
| − | SlicerRT is | + | * SlicerRT is a radiation therapy toolkit for 3D Slicer, containing generic RT features for import/export, analysis, visualization, aiming to make 3D Slicer a powerful radiotherapy research platform. SlicerRT development is currently funded by CANARIE.<br>SlicerRT was originally created via funding by Cancer Care Ontario and the Ontario Consortium for Adaptive Interventions in Radiation Oncology (OCAIRO) to provide free, open-source toolset for radiotherapy and related image-guided interventions. |
| − | |||
| − | The SlicerRT extension incorporates [[Documentation/{{documentation/version}}/Extensions/Plastimatch|Plastimatch]] modules and algorithms. | + | * The SlicerRT extension incorporates [[Documentation/{{documentation/version}}/Extensions/Plastimatch|Plastimatch]] modules and algorithms. |
| + | |||
| + | * Additional information for users can be found on the [[Documentation/SlicerRT/UsersGuide|User's Guide]] page <br> | ||
| | | | ||
|} | |} | ||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Modules}} | {{documentation/{{documentation/version}}/extension-section|Modules}} | ||
| + | {| | ||
| + | | | ||
| − | *[[Documentation/{{documentation/version}}/Modules/ | + | *[[Documentation/{{documentation/version}}/Modules/ExternalBeamPlanning|External Beam Planning]] (Treatment planning) |
| − | * | + | * Dose analysis |
| − | *[[Documentation/{{documentation/version}}/Modules/DoseVolumeHistogram|Dose volume histogram]] | + | **[[Documentation/{{documentation/version}}/Modules/DoseVolumeHistogram|Dose volume histogram]] |
| − | *[[Documentation/{{documentation/version}}/Modules/DoseAccumulation|Dose accumulation]] | + | **[[Documentation/{{documentation/version}}/Modules/DoseAccumulation|Dose accumulation]] |
| − | *[[Documentation/{{documentation/version}}/Modules/DoseComparison|Dose comparison]] | + | **[[Documentation/{{documentation/version}}/Modules/DoseComparison|Dose comparison]] (Gamma dose similarity index) |
| − | *[[Documentation/{{documentation/version}}/Modules/Isodose|Isodose line and surface display]] | + | **[[Documentation/{{documentation/version}}/Modules/Isodose|Isodose line and surface display]] |
| − | *[[Documentation/{{documentation/version}}/Modules/ | + | * Contour analysis |
| − | *[[Documentation/{{documentation/version}}/Modules/ | + | **[[Documentation/{{documentation/version}}/Modules/SegmentComparison|Segment comparison]] (Dice Similarity Coefficient, Hausdorff distances) |
| − | *[[Documentation/{{documentation/version}}/Modules/ | + | **[[Documentation/{{documentation/version}}/Modules/SegmentMorphology|Segment morphology]] (Add/remove margin, Unify, Intersect, etc.) |
| − | *[[Documentation/{{documentation/version}}/Modules/ | + | * I/O |
| + | **[[Documentation/{{documentation/version}}/Modules/DicomRtImport|DICOM-RT import]], [https://www.slicer.org/wiki/Documentation/Nightly/Modules/DICOM#DICOM_export export] (handles datasets of types RT Structure Set, RT Dose, RT Plan, RT Image) | ||
| + | **[[Documentation/{{documentation/version}}/Modules/DicomSroImport|DICOM-SRO import/export]] (handles DICOM Spatial Registration object, both rigid and deformable) | ||
| + | *[https://github.com/SlicerRt/SlicerRT/tree/master/BatchProcessing Batch processing scripts] (currently only one is available for command-line conversion of RTSS to volume nodes) | ||
| − | * Modules from [[Documentation/{{documentation/version}}/Extensions/Plastimatch|Plastimatch]] | + | * Modules from [[Documentation/{{documentation/version}}/Extensions/Plastimatch|Plastimatch]] |
| − | **[[Documentation/{{documentation/version}}/Modules/PlmBSplineDeformableRegistration|Plastimatch Automatic deformable image registration | + | **[[Documentation/{{documentation/version}}/Modules/PlmBSplineDeformableRegistration|Plastimatch Automatic deformable image registration]] |
| − | + | **[[Documentation/{{documentation/version}}/Modules/PlmLANDWARP|Plastimatch LANDWARP Landmark]] [[image:UnderConstruction.png|tumb|10px]] | |
| − | |||
| − | **[[Documentation/{{documentation/version}}/Modules/PlmLANDWARP|Plastimatch LANDWARP Landmark]] | ||
| − | |||
<!-- | <!-- | ||
| − | |||
| − | |||
**[[Documentation/{{documentation/version}}/Modules/PlmSyntheticImageGeneration|Plastimatch Synthetic Image Generation]] (Greg Sharp)[[image:UnderConstruction.png|tumb|10px]] | **[[Documentation/{{documentation/version}}/Modules/PlmSyntheticImageGeneration|Plastimatch Synthetic Image Generation]] (Greg Sharp)[[image:UnderConstruction.png|tumb|10px]] | ||
| − | |||
--> | --> | ||
| + | |||
| + | * Former SlicerRT modules integrated to Slicer core | ||
| + | **[[Documentation/{{documentation/version}}/Modules/Data|Subject hierarchy]] | ||
| + | **[[Documentation/{{documentation/version}}/Modules/Transforms|Transform visualizer]] | ||
| + | **DICOM-RT export, as [[Documentation/Labs/DICOMExport|improved DICOM export function]] | ||
| + | **[[Documentation/{{documentation/version}}/Modules/Segmentations|Segmentations]] | ||
| + | **[[Documentation/{{documentation/version}}/Modules/SegmentEditor|Segment Editor]] | ||
| + | <!-- | ||
| + | * Experimental modules | ||
| + | **[[Documentation/{{documentation/version}}/Modules/ExternalBeamPlanning|External beam planning]] [[image:UnderConstruction.png|tumb|10px]] | ||
| + | --> | ||
| + | | [[File:SlicerRt_Montage.jpg|512px|SlicerRT highlights]] | ||
| + | |} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| + | |||
| + | |||
{{documentation/{{documentation/version}}/extension-section|Use Cases}} | {{documentation/{{documentation/version}}/extension-section|Use Cases}} | ||
* Comparison of dose maps and dose volume histograms from various treatment planning systems | * Comparison of dose maps and dose volume histograms from various treatment planning systems | ||
| − | * Evaluation of the effect of image-based non-rigid patient motion compensation | + | * Evaluation of the effect of different adaptive techniques (IGRT, image-based non-rigid patient motion compensation, etc.) |
| + | ** Calculate couch shift parameters for patient setup correction in IGRT | ||
* Dose accumulation with motion compensation | * Dose accumulation with motion compensation | ||
| + | * Testing of treatment planning algorithms | ||
| + | * Calculation of PTV margin | ||
| + | * Proton dose calculation | ||
| + | * Gel dosimetry analysis | ||
| + | * Tumor volume tracking | ||
| + | * Treatment plan similarity measurement in the cloud | ||
| + | * Batch structure set conversion | ||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Tutorials}} | {{documentation/{{documentation/version}}/extension-section|Tutorials}} | ||
| − | * '''Summer NA-MIC week 2013 tutorial''' | + | |
| + | === Comprehensive tutorials === | ||
| + | |||
| + | * '''Image-guided radiation therapy tutorial 2019''' ''(recommended)'' | ||
| + | ** [https://github.com/SlicerRt/SlicerRtDoc/blob/master/tutorials/SlicerRT_Tutorial_IGRT_4.11.pptx Slides including dataset] | ||
| + | * '''World Congress 2015 tutorial''' | ||
| + | ** Tutorial presentation: [https://www.dropbox.com/s/b7qx3n10s52o5f8/SlicerRT_WorldCongress_TutorialIGRT.pptx?dl=0 pptx] [https://github.com/SlicerRt/SlicerRtDoc/raw/master/tutorials/SlicerRT_WorldCongress_TutorialIGRT.pdf pdf] | ||
| + | ** Dataset: [http://slicer.kitware.com/midas3/download/item/205391/WC2015_Gel_Slicelet_Dataset.zip download] from MIDAS | ||
| + | * '''Summer NA-MIC week 2013 tutorial''' | ||
** Tutorial presentation: [http://wiki.na-mic.org/Wiki/images/b/b0/SlicerRT_TutorialContestSummer2013.pdf download] from Slicer wiki | ** Tutorial presentation: [http://wiki.na-mic.org/Wiki/images/b/b0/SlicerRT_TutorialContestSummer2013.pdf download] from Slicer wiki | ||
** Sample data: [http://slicer.kitware.com/midas3/download/folder/1345/SlicerRtTutorial_Namic2013June.zip download] from MIDAS | ** Sample data: [http://slicer.kitware.com/midas3/download/folder/1345/SlicerRtTutorial_Namic2013June.zip download] from MIDAS | ||
| − | * ECR 2013 - Medical University Vienna workshop | + | * '''ECR 2013 - Medical University Vienna workshop''' |
** Workshop material: [http://www.na-mic.org/Wiki/index.php/File:Pinter_MedUni2013_Workshop.pdf download] from NA-MIC.org | ** Workshop material: [http://www.na-mic.org/Wiki/index.php/File:Pinter_MedUni2013_Workshop.pdf download] from NA-MIC.org | ||
| − | * RSNA 2012 tutorial | + | * '''RSNA 2012 tutorial''' |
| − | ** Tutorial description: [ | + | ** Tutorial description: [http://www.donotlink.com/bEo SlicerRT wiki: Slicer tutorials at RSNA 2012] |
** Sample data: [http://slicer.kitware.com/midas3/folder/859 download] SlicerRT ART dose verification data from Midas server | ** Sample data: [http://slicer.kitware.com/midas3/folder/859 download] SlicerRT ART dose verification data from Midas server | ||
| − | * | + | |
| − | * | + | === Module tutorials === |
| − | * | + | |
| + | * [https://github.com/SlicerRt/SlicerRtDoc/blob/master/tutorials/SlicerRT_Tutorial_OrthovoltageDoseEngine.pptx External beam planning tutorial for orthovoltage RT] (uses EGSnrc) | ||
| + | * [https://github.com/SlicerRt/SlicerRtDoc/blob/master/tutorials/SlicerRT_Tutorial_DoseSurfaceHistogram.pptx Dose surface histogram tutorial] | ||
| + | * [https://github.com/SlicerRt/SlicerRtDoc/blob/master/tutorials/SlicerRT_Tutorial_Isodose.pptx Isodose tutorial] | ||
| + | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Similar Extensions}} | {{documentation/{{documentation/version}}/extension-section|Similar Extensions}} | ||
* [[Documentation/{{documentation/version}}/Extensions/Plastimatch|Plastimatch]]: SlicerRT and Plastimatch are complementary software libraries. Plastimatch focuses on delivering new computational methods radiotherapy, while SlicerRT aims for providing an easy-to-use interface for a wide range of stable, well-tested radiotherapy related features. SlicerRT uses Plastimatch internally for certain operations. | * [[Documentation/{{documentation/version}}/Extensions/Plastimatch|Plastimatch]]: SlicerRT and Plastimatch are complementary software libraries. Plastimatch focuses on delivering new computational methods radiotherapy, while SlicerRT aims for providing an easy-to-use interface for a wide range of stable, well-tested radiotherapy related features. SlicerRT uses Plastimatch internally for certain operations. | ||
| + | * [[Documentation/{{documentation/version}}/Modules/GelDosimetry|Gel Dosimetry]]: Slicelet facilitating a streamlined workflow to perform true 3D gel dosimetry analysis for commissioning linacs and evaluating new dose calculation procedures | ||
| + | * [[Documentation/{{documentation/version}}/Modules/FilmDosimetry|Film Dosimetry]]: Slicelet supporting workflow to perform 2D film dosimetry analysis for commissioning new radiation techniques and to validate the accuracy of radiation treatment by enabling visual comparison of the planned dose to the delivered dose | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|References}} | {{documentation/{{documentation/version}}/extension-section|References}} | ||
| − | + | ==How to cite== | |
| + | Please cite the following paper when referring to SlicerRt in your publication:<br> | ||
| + | C. Pinter, A. Lasso, A. Wang, D. Jaffray and G. Fichtinger, [http://perk.cs.queensu.ca/sites/perk.cs.queensu.ca/files/Pinter2012_0.pdf "SlicerRT – Radiation therapy research toolkit for 3D Slicer"], Med. Phys., 39(10) pp. 6332-6338, 2012 | ||
| + | <pre> | ||
| + | @ARTICLE{Pinter2012, | ||
| + | author = {Pinter, C. and Lasso, A. and Wang, A. and Jaffray, D. and Fichtinger, G.}, | ||
| + | title = {SlicerRT – Radiation therapy research toolkit for 3D Slicer}, | ||
| + | journal = {Med. Phys.}, | ||
| + | year = {2012}, | ||
| + | volume = {39}, | ||
| + | number = {10}, | ||
| + | pages = {6332-6338}, | ||
| + | } | ||
| + | </pre> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Information for Developers}} | {{documentation/{{documentation/version}}/extension-section|Information for Developers}} | ||
| + | * [https://github.com/SlicerRt/SlicerRT/wiki/SlicerRt-developers-page SlicerRT developers wiki page] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-footer}} | {{documentation/{{documentation/version}}/extension-footer}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 02:26, 27 November 2019
Home < Documentation < Nightly < Extensions < SlicerRT
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
Authors: Csaba Pinter (PerkLab, Queen's University), Andras Lasso (PerkLab, Queen's University)
Contributors: Greg Sharp (Massachusetts General Hospital), Kevin Wang (Princess Margaret Hospital, UHN Toronto), Steve Pieper (Isomics)
Contacts:
- Csaba Pinter, <email>pinter.csaba@gmail.com</email>
- Andras Lasso, <email>lasso@cs.queensu.ca</email>
- Slicer Forum
- How to report an error
Website: slicerrt.org
License: Slicer license
Download/install: install 3D Slicer, start 3D Slicer, open the Extension Manager, install the SlicerRT extension (see more details on the download page)
Extension Description
|
Modules
|
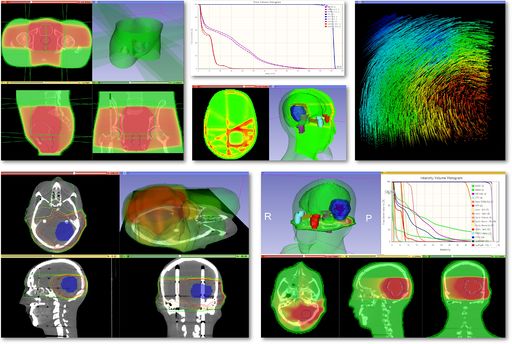
|
Use Cases
- Comparison of dose maps and dose volume histograms from various treatment planning systems
- Evaluation of the effect of different adaptive techniques (IGRT, image-based non-rigid patient motion compensation, etc.)
- Calculate couch shift parameters for patient setup correction in IGRT
- Dose accumulation with motion compensation
- Testing of treatment planning algorithms
- Calculation of PTV margin
- Proton dose calculation
- Gel dosimetry analysis
- Tumor volume tracking
- Treatment plan similarity measurement in the cloud
- Batch structure set conversion
Tutorials
Comprehensive tutorials
- Image-guided radiation therapy tutorial 2019 (recommended)
- World Congress 2015 tutorial
- Summer NA-MIC week 2013 tutorial
- ECR 2013 - Medical University Vienna workshop
- Workshop material: download from NA-MIC.org
- RSNA 2012 tutorial
- Tutorial description: SlicerRT wiki: Slicer tutorials at RSNA 2012
- Sample data: download SlicerRT ART dose verification data from Midas server
Module tutorials
- External beam planning tutorial for orthovoltage RT (uses EGSnrc)
- Dose surface histogram tutorial
- Isodose tutorial
Similar Extensions
- Plastimatch: SlicerRT and Plastimatch are complementary software libraries. Plastimatch focuses on delivering new computational methods radiotherapy, while SlicerRT aims for providing an easy-to-use interface for a wide range of stable, well-tested radiotherapy related features. SlicerRT uses Plastimatch internally for certain operations.
- Gel Dosimetry: Slicelet facilitating a streamlined workflow to perform true 3D gel dosimetry analysis for commissioning linacs and evaluating new dose calculation procedures
- Film Dosimetry: Slicelet supporting workflow to perform 2D film dosimetry analysis for commissioning new radiation techniques and to validate the accuracy of radiation treatment by enabling visual comparison of the planned dose to the delivered dose
References
How to cite
Please cite the following paper when referring to SlicerRt in your publication:
C. Pinter, A. Lasso, A. Wang, D. Jaffray and G. Fichtinger, "SlicerRT – Radiation therapy research toolkit for 3D Slicer", Med. Phys., 39(10) pp. 6332-6338, 2012
@ARTICLE{Pinter2012,
author = {Pinter, C. and Lasso, A. and Wang, A. and Jaffray, D. and Fichtinger, G.},
title = {SlicerRT – Radiation therapy research toolkit for 3D Slicer},
journal = {Med. Phys.},
year = {2012},
volume = {39},
number = {10},
pages = {6332-6338},
}




