Difference between revisions of "Documentation/4.2/Training"
From Slicer Wiki
Tag: 2017 source edit |
|||
| (28 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | <noinclude>{{documentation/ | + | <noinclude>{{documentation/historicaltraining}}</noinclude> |
__TOC__ | __TOC__ | ||
| Line 108: | Line 108: | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | *[ | + | *[[Media:Cardiac MRI Toolkit Tutorial Summer2013.pdf|Cardiac MRI Toolkit]] |
*Authors: Salma Bengali, Josh Cates, SCI, Utah | *Authors: Salma Bengali, Josh Cates, SCI, Utah | ||
| − | *Dataset: [ | + | *Dataset: [[Media:Cardiac_MRI_Toolkit_Tutorial_Data.zip|Cardiac MRI Toolkit Tutorial Dataset]] |
| + | |align="right"| | ||
| + | [[Image:CMRToolkit_Tutorial_Image.png|250px]] | ||
| + | |} | ||
==HelloCLI== | ==HelloCLI== | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | *[ | + | *[[Media:Hello_CLI_TutorialContestSummer2013.pdf|HelloCLI]] |
*Authors: Nadya Shusharina, Greg Sharp, MGH, Boston | *Authors: Nadya Shusharina, Greg Sharp, MGH, Boston | ||
| − | *Dataset: [ | + | *Dataset: [[Media:Hello_CLI_TutorialContestSummer2013.zip|HelloCLI Dataset]] |
| + | |align="right"| | ||
| + | [[Image:Cli_icon.png|300px]] | ||
| + | |} | ||
==SlicerRT== | ==SlicerRT== | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | *[ | + | *[[Media:SlicerRT_TutorialContestSummer2013.pdf|SlicerRT Tutorial]] |
| − | *Authors: Csaba Pinter, Andras Lasso(Queen's), Kevin Wang(PMH, Toronto) | + | *Authors: Csaba Pinter, Andras Lasso (Queen's), Kevin Wang (PMH, Toronto) |
| − | *Dataset: [ | + | *Dataset: [[Media:CsabaPinter-SlicerRtTutorial_Namic2013June.zip|SlicerRT Dataset]] |
| + | |align="right"| | ||
| + | [[Image:667px-SlicerRT_0.10_IsocenterShiftingEvaluation.png|250px]] | ||
| + | |} | ||
| + | |||
| + | ==DTIPrep== | ||
| + | {|width="100%" | ||
| + | | | ||
| + | *[[Media:DTIPrep_TutorialContestSummer2013.pdf|DTIPrep]] | ||
| + | *Authors: Dave Welch, SINAPSE, IOWA | ||
| + | *Dataset: [[Media:DTIPrepData_TutorialContestSummer2013.zip|DTIPrep Dataset]] | ||
=Summer 2012 Tutorial contest= | =Summer 2012 Tutorial contest= | ||
| Line 168: | Line 184: | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | *[http:// | + | *[http://www.slicer.org/w/img_auth.php/f/f1/OpenIGTLinkTutorial_Slicer4.1.0_JunichiTokuda_Apr2012.pdf OpenIGTLink] |
*Authors: Junichi Tokuda, BWH | *Authors: Junichi Tokuda, BWH | ||
|align="right"| | |align="right"| | ||
| Line 181: | Line 197: | ||
* Author: Justin Kirby | * Author: Justin Kirby | ||
*Audience: Novice 3D Slicer users interested in visualizing prostate segmentations hosted on TCIA. | *Audience: Novice 3D Slicer users interested in visualizing prostate segmentations hosted on TCIA. | ||
| − | *Summary: 3D Slicer was used to generate NRRD segmentations of the following prostate components: prostate gland boundary; internal capsule; central gland, peripheral zone; seminal vesicles; urethra; cancer – dominant nodule; neurovascular bundle; penile bulb; ejaculatory duct; veru-montanum; rectum. These segmentations are currently available for [https://wiki.cancerimagingarchive.net/display/Public/Prostate-Diagnosis 5 subjects from TCIA’s Prostate-Diagnosis image collection]. The [http://www.slicer.org/ | + | *Summary: 3D Slicer was used to generate NRRD segmentations of the following prostate components: prostate gland boundary; internal capsule; central gland, peripheral zone; seminal vesicles; urethra; cancer – dominant nodule; neurovascular bundle; penile bulb; ejaculatory duct; veru-montanum; rectum. These segmentations are currently available for [https://wiki.cancerimagingarchive.net/display/Public/Prostate-Diagnosis 5 subjects from TCIA’s Prostate-Diagnosis image collection]. The [http://www.slicer.org/wiki/File:ProstateDx-01-0006-1.zip archive] contains both the DICOM images and the NRRD file for one of the 5 available subjects. |
|align="right"|[[Image:NCIA-prostate.png|250px]] | |align="right"|[[Image:NCIA-prostate.png|250px]] | ||
|- | |- | ||
Latest revision as of 22:13, 22 November 2022
Home < Documentation < 4.2 < Training
 |
This section is currently out-of-date and may contain errors but is retained for historical reference. Up-to-date training materials can be found at Documentation/Nightly/Training |
Contents
Introduction: Slicer 4.2 Tutorials
- This page contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version 4.2 release) to accomplish certain tasks.
- For tutorials for other versions of Slicer, please visit the Slicer training portal.
- For "reference manual" style documentation, please visit the Slicer 4.2 documentation page.
- For an overview of Slicer and its abilities please see 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network.
- For questions related to the Slicer4 Compendium, please send an e-mail to Sonia Pujol, Ph.D..
General Introduction
Slicer Welcome Tutorial
|
Slicer4Minute Tutorial
|
Slicer4 Data Loading and 3D Visualization
|
Tutorials for software developers
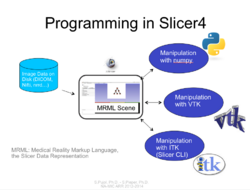
Slicer4 Programming Tutorial
|
Specific functions
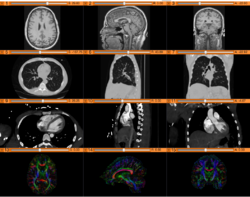
Slicer4 Diffusion Tensor Imaging Tutorial
|
Slicer4 Neurosurgical Planning Tutorial
|
Slicer4 3D Visualization of DICOM images for Radiology Applications
|
Slicer4 Quantitative Imaging tutorial
|
Summer 2013 Tutorial contest
Cardiac MRI Toolkit
|
HelloCLI
|
SlicerRT
|
DTIPrep
Summer 2012 Tutorial contestAutomatic Left Atrial Scar Segmenter
Qualitative and quantitative comparison of two RT dose distributions
Dose accumulation for adaptive radiation therapy
WebGL Export
OpenIGTLink
Additional resources
External ResourcesUsing the EditorThis set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field:
User ContributionsSee the User Contributions Page for more content. |