Difference between revisions of "Documentation/Nightly/Modules/AutomatedLASegmentation"
(Added documentation for graph cut module) |
m |
||
| (6 intermediate revisions by 3 users not shown) | |||
| Line 8: | Line 8: | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | Author: Salma Bengali, CARMA Center, University of Utah<br> | + | Author: Umme Salma Bengali, CARMA Center, University of Utah<br> |
Contributors: Gopal Veni, Alan Morris, Josh Cates, Rob MacLeod, Ross Whitaker, CARMA Center, University of Utah<br> | Contributors: Gopal Veni, Alan Morris, Josh Cates, Rob MacLeod, Ross Whitaker, CARMA Center, University of Utah<br> | ||
| − | Contact: Salma Bengali: <email>salma.bengali@carma.utah.edu</email> <br> | + | Contact: Umme Salma Bengali: <email>salma.bengali@carma.utah.edu</email> <br> |
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | {| | + | {{documentation/{{documentation/version}}/module-introduction-logo-gallery |
| − | + | |{{collaborator|logo|uofu}}|{{collaborator|longname|uofu}} | |
| − | + | }} | |
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | This module automatically segments the left atrium from a cardiac LGE-MRI image volume. The input to the module is a cardiac LGE-MRI image, a smoothness parameter and model data. The model is arbitrarily selected and can be changed to check if it improves the segmentation output. | + | '''Note: This module is licensed for research purposes only. This module relies on code made available by Vladimir Kolmogorov for research use only. See [http://vision.csd.uwo.ca/code/ this web site] for more information.''' |
| + | |||
| + | The module automatically segments the left atrium from a cardiac LGE-MRI image volume. The input to the module is a cardiac LGE-MRI image, a smoothness parameter and model data. The model is arbitrarily selected and can be changed to check if it improves the segmentation output. | ||
Model data to run this module can be downloaded from the [http://slicer.kitware.com/midas3/download/folder/1550/LA+Segmentation+using+Graph+Cuts.zip Slicer Midas repository]. | Model data to run this module can be downloaded from the [http://slicer.kitware.com/midas3/download/folder/1550/LA+Segmentation+using+Graph+Cuts.zip Slicer Midas repository]. | ||
<!-- | <!-- | ||
| Line 62: | Line 64: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | + | *Boykov, Yuri, and Vladimir Kolmogorov. "An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision." Pattern Analysis and Machine Intelligence, IEEE Transactions on 26, no. 9 (2004): 1124-1137. | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 02:44, 12 August 2015
Home < Documentation < Nightly < Modules < AutomatedLASegmentation
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Author: Umme Salma Bengali, CARMA Center, University of Utah | |||
|
Module Description
Note: This module is licensed for research purposes only. This module relies on code made available by Vladimir Kolmogorov for research use only. See this web site for more information.
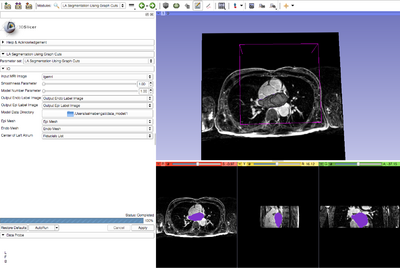
The module automatically segments the left atrium from a cardiac LGE-MRI image volume. The input to the module is a cardiac LGE-MRI image, a smoothness parameter and model data. The model is arbitrarily selected and can be changed to check if it improves the segmentation output. Model data to run this module can be downloaded from the Slicer Midas repository.
Use Cases
This module can be used to automatically segment the Left Atrium from a cardiac LGE-MRI image.
Tutorials
[[Media:]]
Panels and their use
- Input Image : Select the cardiac LGE-MRI image volume
- Smoothness Parameter : Select the smoothness parameter value
- Model Number: Select the model number
- Output Endo Label Image: Create the output endo label image
- Output Epi Label Image: Create the output epi label image
- Model Data Directory: Select the directory of the input model
- Epi Mesh: Create the output epi mesh
- Endo Mesh: Create the output endo mesh
- Center of Left Atrium: Select the approximate center of the left atrium by placing a fiducial
Similar Modules
N/A
References
- Boykov, Yuri, and Vladimir Kolmogorov. "An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision." Pattern Analysis and Machine Intelligence, IEEE Transactions on 26, no. 9 (2004): 1124-1137.
Information for Developers
The source code for this module can be found at the CARMA GitHub repository.