Difference between revisions of "Modules:Modelmaker-Documentation"
| (4 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
[[Documentation|Return to Slicer Documentation]] | [[Documentation|Return to Slicer Documentation]] | ||
| − | [[Image:ModelMakerModule.png|thumb|right|The Model Maker Module]] | + | [[Image:ModelMakerModule.png|thumb|350px|right|The Model Maker Module]] |
| − | + | ||
| − | + | The Modelmaker is used to create 3D surface models from segmented image data, called label maps. Label maps can be the result of automated segmentation or interactive editing. | |
| − | *Models are imported into Slicer under a model hierarchy node, and their colors are set by the color table associated with the input volume. | + | |
| − | *Create Multiple: | + | The model maker is a pipeline of algorithms that start from the input label map, creates a binary label map with just the label(s) of interest set to 1, everything else to 0, generates a marching cubes model, runs triangle reduction and triangle smoothing algorithms. The pipeline was optimized for 1mm brain MRI data. For other geometries, adjustments of the parameters might be necessary. |
| − | + | ||
| + | The model maker is compiled as both a command line executable passed arguments via the command line, and as a binary plug in that communicates through shared memory with Slicer3. The automatically created GUI, found by clickign on the Modules menu, Model Generation category, has the following elements: | ||
| + | |||
| + | * Input/Output: | ||
| + | **The Input Volume drop down menu is populated with the label map volumes that are present in the scene | ||
| + | **Models are imported into Slicer under a model hierarchy node, and their colors are set by the color table associated with the input label map volume. The model hierarchy node '''must be created''' before running the model maker, by selecting ''Create New ModelHierarchy'' from the Models drop down menu. | ||
| + | *Create Multiple: | ||
| + | **Any text entered in the ''Model Prefix'' box will be the starting string for the created model's file names. The label number and the color name will also be part of the file name. | ||
| + | **If you want to create all models that correspond to all values in a labelmap volume, click the ''Generate All Models'' and ''Joint Smoothing'' boxes | ||
| + | **''Joint Smoothing'' will ensure that all resulting models fit together smoothly, like jigsaw puzzle pieces. Otherwise the models will be smoothed independently and may overlap. | ||
| + | **If you specify a list of Labels, it will override any start/end label settings. If you click ''Generate All Models'' it will override the list of labels and any start/end label settings. | ||
*Model Maker Parameters: | *Model Maker Parameters: | ||
| − | **You can set the number of smoothing iterations, target reduction in number of polygons (decimal percentage). Use 0 and 1 if you wish no smoothing nor decimation. | + | **You can set the number of smoothing iterations, target reduction in number of polygons (decimal percentage). Use 0 and 1 if you wish no smoothing nor decimation. |
| + | **You can control the type of smoothing done on the models by selecting a ''Filter Type''. | ||
**You can set the flags to split normals or generate point normals in this pane as well. | **You can set the flags to split normals or generate point normals in this pane as well. | ||
| − | **You can save a copy of the models after intermediate steps (marching cubes, smoothing, and decimation if not joint smoothing, otherwise just after decimation); these models are not saved in the mrml file, turn off deleting temporary files first in the tcl window: [$::slicer3::CommandLineModuleGUI_Model_Maker GetLogic] DeleteTemporaryFilesOff | + | ***''Split Normals'' is useful for visualizing sharp features. However it creates holes in surfaces which affects measurements. |
| − | *Anatomical Information: Here you can specify an external file linking label values with anatomical names. | + | ***''Point Normals'' is turned on if you wish to calculate the normal vectors for the points. |
| − | This was the original method of linking label map values to resultant model colors and names, but it has been | + | **You can save a copy of the models after intermediate steps (marching cubes, smoothing, and decimation if not joint smoothing, otherwise just after decimation); these models are not saved in the mrml file, turn off deleting temporary files first in the tcl window, and then load them manually: |
| + | [$::slicer3::CommandLineModuleGUI_Model_Maker GetLogic] DeleteTemporaryFilesOff | ||
| + | *Anatomical Information: | ||
| + | **Here you can specify an external file linking label values with anatomical names. | ||
| + | This was the original method of linking label map values to resultant model colors and names, but it has been superseded by an internal linkage between the input label map volume and it's color node. If you wish to specify a file for a volume node that does not have a color node associated with it. It must be a comma separated file, extension .csv, in the format: | ||
anatomyName1,label1 | anatomyName1,label1 | ||
anatomyName2,label2 | anatomyName2,label2 | ||
where anatomyName is a string, and label is an integer. If the volume node has a color node associated with it, that will override the anatomy label file. | where anatomyName is a string, and label is an integer. If the volume node has a color node associated with it, that will override the anatomy label file. | ||
| + | |||
| + | *You can choose to not generate models from labels that do not have names. | ||
| + | |||
| + | |||
| + | == Trouble shooting == | ||
| + | |||
| + | |||
| + | The two main reasons that the Model Maker may fail are: | ||
| + | # the models were not created (see the error log; are you setting the Model Hierarchy drop down to create a new one?) | ||
| + | # the models don't load/display. | ||
| + | |||
| + | If you run the Model Maker and "nothing happens" here are some ways to track down the problem. | ||
| + | |||
| + | * Check the Window->Error Log to see if there was a problem recorded after the Model Maker returned | ||
| + | * Check in your temporary directory (View->Application Settings->Module Settings) to see if the .vtk files were created. If the files are there, try loading them manually via the Add Data panel. | ||
| + | * There should also be a mrml file in the Temporary Directory that points to the models files and also defines the model colours, try loading that manually, using File->Load Scene. | ||
| + | * There may be a bug in the rendering pipeline, check the Data module to see if the models are listed there. If so, try rotating the scene or adding a fiducial point to trigger a render. | ||
Latest revision as of 18:02, 17 December 2008
Home < Modules:Modelmaker-DocumentationReturn to Slicer Documentation
The Modelmaker is used to create 3D surface models from segmented image data, called label maps. Label maps can be the result of automated segmentation or interactive editing.
The model maker is a pipeline of algorithms that start from the input label map, creates a binary label map with just the label(s) of interest set to 1, everything else to 0, generates a marching cubes model, runs triangle reduction and triangle smoothing algorithms. The pipeline was optimized for 1mm brain MRI data. For other geometries, adjustments of the parameters might be necessary.
The model maker is compiled as both a command line executable passed arguments via the command line, and as a binary plug in that communicates through shared memory with Slicer3. The automatically created GUI, found by clickign on the Modules menu, Model Generation category, has the following elements:
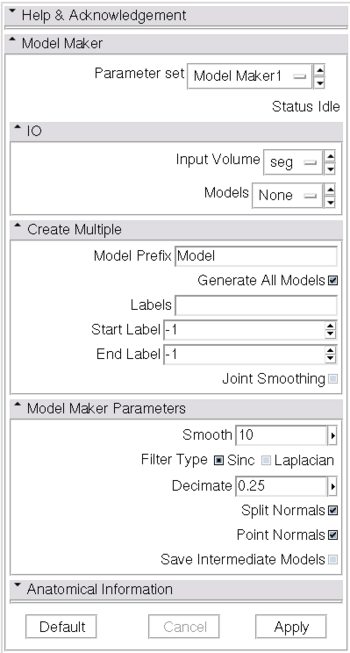
- Input/Output:
- The Input Volume drop down menu is populated with the label map volumes that are present in the scene
- Models are imported into Slicer under a model hierarchy node, and their colors are set by the color table associated with the input label map volume. The model hierarchy node must be created before running the model maker, by selecting Create New ModelHierarchy from the Models drop down menu.
- Create Multiple:
- Any text entered in the Model Prefix box will be the starting string for the created model's file names. The label number and the color name will also be part of the file name.
- If you want to create all models that correspond to all values in a labelmap volume, click the Generate All Models and Joint Smoothing boxes
- Joint Smoothing will ensure that all resulting models fit together smoothly, like jigsaw puzzle pieces. Otherwise the models will be smoothed independently and may overlap.
- If you specify a list of Labels, it will override any start/end label settings. If you click Generate All Models it will override the list of labels and any start/end label settings.
- Model Maker Parameters:
- You can set the number of smoothing iterations, target reduction in number of polygons (decimal percentage). Use 0 and 1 if you wish no smoothing nor decimation.
- You can control the type of smoothing done on the models by selecting a Filter Type.
- You can set the flags to split normals or generate point normals in this pane as well.
- Split Normals is useful for visualizing sharp features. However it creates holes in surfaces which affects measurements.
- Point Normals is turned on if you wish to calculate the normal vectors for the points.
- You can save a copy of the models after intermediate steps (marching cubes, smoothing, and decimation if not joint smoothing, otherwise just after decimation); these models are not saved in the mrml file, turn off deleting temporary files first in the tcl window, and then load them manually:
[$::slicer3::CommandLineModuleGUI_Model_Maker GetLogic] DeleteTemporaryFilesOff
- Anatomical Information:
- Here you can specify an external file linking label values with anatomical names.
This was the original method of linking label map values to resultant model colors and names, but it has been superseded by an internal linkage between the input label map volume and it's color node. If you wish to specify a file for a volume node that does not have a color node associated with it. It must be a comma separated file, extension .csv, in the format:
anatomyName1,label1 anatomyName2,label2
where anatomyName is a string, and label is an integer. If the volume node has a color node associated with it, that will override the anatomy label file.
- You can choose to not generate models from labels that do not have names.
Trouble shooting
The two main reasons that the Model Maker may fail are:
- the models were not created (see the error log; are you setting the Model Hierarchy drop down to create a new one?)
- the models don't load/display.
If you run the Model Maker and "nothing happens" here are some ways to track down the problem.
- Check the Window->Error Log to see if there was a problem recorded after the Model Maker returned
- Check in your temporary directory (View->Application Settings->Module Settings) to see if the .vtk files were created. If the files are there, try loading them manually via the Add Data panel.
- There should also be a mrml file in the Temporary Directory that points to the models files and also defines the model colours, try loading that manually, using File->Load Scene.
- There may be a bug in the rendering pipeline, check the Data module to see if the models are listed there. If so, try rotating the scene or adding a fiducial point to trigger a render.