Difference between revisions of "Documentation/4.2/Extensions/SlicerRT"
m |
|||
| (8 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <noinclude>{{documentation/versioncheck}}</noinclude> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
| Line 4: | Line 5: | ||
[[File:LogoCco.png|Cancer Care Ontario]] | [[File:LogoCco.png|Cancer Care Ontario]] | ||
[[File:Logo-SparKit.png|SparKit]] | [[File:Logo-SparKit.png|SparKit]] | ||
| + | [[File:LogoOCAIRO.jpg|SparKit]] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | {{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | ||
| − | Authors: Csaba Pinter (PerkLab, Queen's University), Andras Lasso (PerkLab, Queen's University), Kevin Wang (Radiation Medicine Program, Princess Margaret Hospital, University Health Network Toronto)<br> | + | Authors: <b>Csaba Pinter</b> (PerkLab, Queen's University), <b>Andras Lasso</b> (PerkLab, Queen's University), <b>Kevin Wang</b> (Radiation Medicine Program, Princess Margaret Hospital, University Health Network Toronto)<br> |
Contributors: Greg Sharp (Massachusetts General Hospital), Steve Pieper (Isomics)<br> | Contributors: Greg Sharp (Massachusetts General Hospital), Steve Pieper (Isomics)<br> | ||
| − | + | Contacts: | |
| − | Website: http:// | + | * Andras Lasso, <email>lasso@cs.queensu.ca</email> |
| + | * Mailing list, <email>slicerrt@alerts.assembla.com</email> (need to register to [http://www.assembla.com Assembla] using the email address from which the email is sent) | ||
| + | * [https://www.assembla.com/spaces/slicerrt/wiki/How_to_report_an_error How to report an error] | ||
| + | Website: [http://slicerrt.github.io slicerrt.org]<br> | ||
License: [http://www.slicer.org/pages/LicenseText Slicer license] | License: [http://www.slicer.org/pages/LicenseText Slicer license] | ||
| Line 37: | Line 42: | ||
*[[Documentation/{{documentation/version}}/Modules/ContourMorphology|Contour morphology]] | *[[Documentation/{{documentation/version}}/Modules/ContourMorphology|Contour morphology]] | ||
<!--*[[Documentation/{{documentation/version}}/Modules/PatientHierarchy|Patient hierarchy]]--> | <!--*[[Documentation/{{documentation/version}}/Modules/PatientHierarchy|Patient hierarchy]]--> | ||
| − | *[[Documentation/{{documentation/version}}/Modules/DeformationFieldVisualizer| | + | *[[Documentation/{{documentation/version}}/Modules/DeformationFieldVisualizer|Transform visualizer (moved to separate extension)]] |
* Modules from [[Documentation/{{documentation/version}}/Extensions/Plastimatch|Plastimatch]] (Greg Sharp) | * Modules from [[Documentation/{{documentation/version}}/Extensions/Plastimatch|Plastimatch]] (Greg Sharp) | ||
| Line 66: | Line 71: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/extension-section|Tutorials}} | {{documentation/{{documentation/version}}/extension-section|Tutorials}} | ||
| − | * Summer NA-MIC week | + | * Summer NA-MIC week 2013 tutorial |
| − | **Tutorial presentation: [ | + | ** Tutorial presentation: [http://wiki.na-mic.org/Wiki/images/b/b0/SlicerRT_TutorialContestSummer2013.pdf download] from Slicer wiki |
| − | ** Sample data: [ | + | ** Sample data: [http://slicer.kitware.com/midas3/download/folder/1345/SlicerRtTutorial_Namic2013June.zip download] from MIDAS |
| + | * ECR 2013 - Medical University Vienna workshop | ||
| + | ** Workshop material: [http://www.na-mic.org/Wiki/index.php/File:Pinter_MedUni2013_Workshop.pdf download] from NA-MIC.org | ||
* RSNA 2012 tutorial | * RSNA 2012 tutorial | ||
** Tutorial description: [https://www.assembla.com/spaces/slicerrt/wiki/20121127_Slicer_tutorials_at_RSNA_2012 SlicerRT wiki: Slicer tutorials at RSNA 2012] | ** Tutorial description: [https://www.assembla.com/spaces/slicerrt/wiki/20121127_Slicer_tutorials_at_RSNA_2012 SlicerRT wiki: Slicer tutorials at RSNA 2012] | ||
** Sample data: [http://slicer.kitware.com/midas3/folder/859 download] SlicerRT ART dose verification data from Midas server | ** Sample data: [http://slicer.kitware.com/midas3/folder/859 download] SlicerRT ART dose verification data from Midas server | ||
| − | * | + | * Summer NA-MIC week 2012 tutorial |
| − | ** | + | ** Tutorial presentation: [https://www.assembla.com/spaces/slicerrt/documents/aAuMq81Eqr4ztvacwqjQXA/download/aAuMq81Eqr4ztvacwqjQXA download] from the SlicerRT website |
| + | ** Sample data: [https://www.assembla.com/spaces/slicerrt/documents/bMuwgYTKur4yP-acwqjQWU/download/bMuwgYTKur4yP-acwqjQWU download] from the SlicerRT website | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 16:07, 21 August 2013
Home < Documentation < 4.2 < Extensions < SlicerRT
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
Authors: Csaba Pinter (PerkLab, Queen's University), Andras Lasso (PerkLab, Queen's University), Kevin Wang (Radiation Medicine Program, Princess Margaret Hospital, University Health Network Toronto)
Contributors: Greg Sharp (Massachusetts General Hospital), Steve Pieper (Isomics)
Contacts:
- Andras Lasso, <email>lasso@cs.queensu.ca</email>
- Mailing list, <email>slicerrt@alerts.assembla.com</email> (need to register to Assembla using the email address from which the email is sent)
- How to report an error
Website: slicerrt.org
License: Slicer license
Extension Description
|
SlicerRT is one of the themes of the SparKit (Software Platform and Adaptive Radiotherapy Kit) project with the goal of making 3D Slicer a powerful radiotherapy research platform. SparKit is a project funded by Cancer Care Ontarioand the Ontario Consortium for Adaptive Interventions in Radiation Oncology (OCAIRO) to provide free, open-source toolset for radiotherapy and related image-guided interventions. The SlicerRT extension incorporates Plastimatch modules and algorithms. |
Modules
- DICOM-RT import
- Contours
- Dose volume histogram
- Dose accumulation
- Dose comparison
- Isodose line and surface display
- Contour comparison
- Contour morphology
- Transform visualizer (moved to separate extension)
- Modules from Plastimatch (Greg Sharp)
- Plastimatch Automatic deformable image registration (Greg Sharp)
- Plastimatch DICOM-RT import (Greg Sharp)
- Plastimatch DICOM-RT export (Greg Sharp)

- Plastimatch LANDWARP Landmark (Greg Sharp)
- Plastimatch XFORMWARP (Greg Sharp)

Important note: BSpline Deformable registration and LANDWARP modules do not show up for Slicer 4.2 on Mac
Use Cases
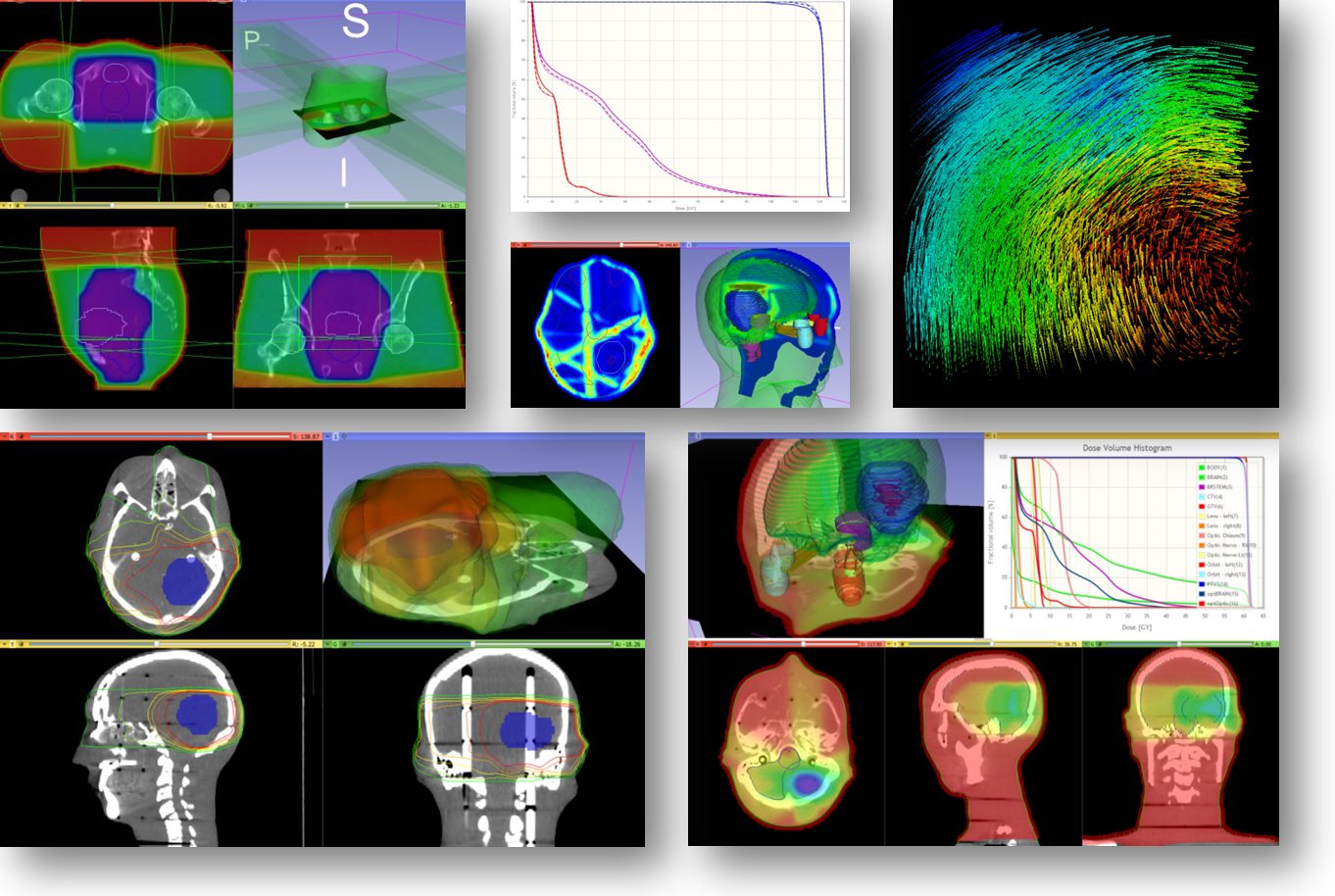
- Comparison of dose maps and dose volume histograms from various treatment planning systems
- Evaluation of the effect of image-based non-rigid patient motion compensation
- Dose accumulation with motion compensation
|
Tutorials
- Summer NA-MIC week 2013 tutorial
- ECR 2013 - Medical University Vienna workshop
- Workshop material: download from NA-MIC.org
- RSNA 2012 tutorial
- Tutorial description: SlicerRT wiki: Slicer tutorials at RSNA 2012
- Sample data: download SlicerRT ART dose verification data from Midas server
- Summer NA-MIC week 2012 tutorial
Similar Extensions
- Plastimatch: SlicerRT and Plastimatch are complementary software libraries. Plastimatch focuses on delivering new computational methods radiotherapy, while SlicerRT aims for providing an easy-to-use interface for a wide range of stable, well-tested radiotherapy related features. SlicerRT uses Plastimatch internally for certain operations.
References
Information for Developers
| Section under construction. |



