Difference between revisions of "Documentation/Nightly/Modules/SlicerModuleIsolatedConnected"
m |
|||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <noinclude>{{documentation/versioncheck}}</noinclude> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
| Line 7: | Line 8: | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | Author: Salma Bengali, CARMA Center, University of Utah<br> | + | Author: Umme Salma Bengali, CARMA Center, University of Utah<br> |
Contributors: Alan Morris, Josh Cates, Rob MacLeod, CARMA Center, University of Utah<br> | Contributors: Alan Morris, Josh Cates, Rob MacLeod, CARMA Center, University of Utah<br> | ||
| − | Contacts: Salma Bengali: <email>salma.bengali@carma.utah.edu</email> <br> | + | Contacts: Umme Salma Bengali: <email>salma.bengali@carma.utah.edu</email> <br> |
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | {| | + | {{documentation/{{documentation/version}}/module-introduction-logo-gallery |
| − | + | |{{collaborator|logo|uofu}}|{{collaborator|longname|uofu}} | |
| − | + | }} | |
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| Line 31: | Line 32: | ||
This module is meant to be used for segmenting the entire endocardium region from a cardiac MRI or MRA image. It can also be used for segmentation of structures in other types of images. | This module is meant to be used for segmenting the entire endocardium region from a cardiac MRI or MRA image. It can also be used for segmentation of structures in other types of images. | ||
{| | {| | ||
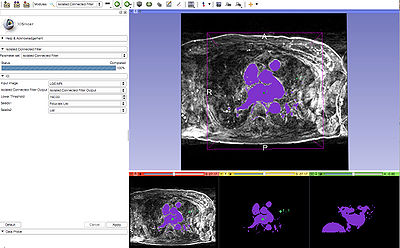
| − | |[[Image:isolatedConnected.jpg|thumb|400px|Left: | + | |[[Image:isolatedConnected.jpg|thumb|400px|Bottom Left:Segmentation overlaid on LGE-MRI image, Center: Segmentation]] |
|} | |} | ||
Latest revision as of 02:44, 12 August 2015
Home < Documentation < Nightly < Modules < SlicerModuleIsolatedConnected
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Author: Umme Salma Bengali, CARMA Center, University of Utah | |||
|
Module Description
This module uses two seed points to segment an image. One seed point (fiducial) is placed within the region to be segmented, and the other is placed outside that region. Multiple seed points can be used as well. The lower threshold is given as an input by the user. The isolated connected image filter will find the isolating upper threshold. If no isolating threshold can be found then the segmentation will fail.
Use Cases
This module is meant to be used for segmenting the entire endocardium region from a cardiac MRI or MRA image. It can also be used for segmentation of structures in other types of images.
Tutorials
Isolated Connected Filter Tutorial
Panels and their use
- Input image
- Output image
- Lower threshold
- Seed1: User placed fiducial/s inside the region to be segmented
- Seed2: User placed fiducial/s outside the region to be segmented
Similar Modules
N/A
References
N/A
Information for Developers
The source code for this module can be found at the CARMA GitHub repository.