Difference between revisions of "Documentation/Nightly/Modules/SampleData"
From Slicer Wiki
(4.1 -> Nightly) |
m (Text replacement - "https?:\/\/(?:www|wiki)\.slicer\.org\/slicerWiki\/index\.php\/([^ ]+) " to "https://www.slicer.org/wiki/$1 ") |
||
| (5 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <noinclude>{{documentation/versioncheck}}</noinclude> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
| Line 7: | Line 8: | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}} }} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}} }} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | + | :'''Author(s)/Contributor(s):''' Steve Pieper (Isomics Inc.), Jean-Christophe Fillion-Robin (Kitware), Julien Finet (Kitware), Benjamin Long (Kitware)<br> | |
| − | + | : '''Acknowledgements:''' This work is part of the [http://www.na-mic.org/ National Alliance for Medical Image Computing] (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149.<br> | |
| − | Contact: Steve Pieper, <email>pieper@bwh.harvard.edu</email><br> | + | : '''Contact:''' Steve Pieper, <email>pieper@bwh.harvard.edu</email><br> |
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
|{{collaborator|logo|isomics}}|{{collaborator|longname|isomics}} | |{{collaborator|logo|isomics}}|{{collaborator|longname|isomics}} | ||
| + | |{{collaborator|logo|kitware}}|{{collaborator|longname|kitware}} | ||
|{{collaborator|logo|namic}}|{{collaborator|longname|namic}} | |{{collaborator|logo|namic}}|{{collaborator|longname|namic}} | ||
}} | }} | ||
| Line 20: | Line 22: | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | + | Provides an easy to download a variety of anonymized datasets (Mr, CT, ...) for learning and testing in Slicer. Because the datasets are hosted in a remote repository, a network connection is required to access them. | |
[[image:Selection 153.png|Sample Data Module]] | [[image:Selection 153.png|Sample Data Module]] | ||
| Line 41: | Line 43: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| − | * The DICOM module can be used to | + | * The DICOM module can be used to access sample data on DICOM servers. |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 50: | Line 52: | ||
This module is written in python, and relies on MRML's ability to access datasets via the http protocol. | This module is written in python, and relies on MRML's ability to access datasets via the http protocol. | ||
| − | The sample data is accessed from the wiki at this url: | + | The sample data is accessed from the wiki at this url: https://www.slicer.org/wiki/SampleData |
| + | Modules can register their own sample data by following [https://github.com/Slicer/Slicer/blob/master-43/Modules/Scripted/SampleData/SampleData.py#L163-182 the example in the module]. | ||
| + | |||
{{documentation/{{documentation/version}}/module-developerinfo}} | {{documentation/{{documentation/version}}/module-developerinfo}} | ||
Latest revision as of 13:21, 27 November 2019
Home < Documentation < Nightly < Modules < SampleData
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
| |||||||
|
Module Description
Provides an easy to download a variety of anonymized datasets (Mr, CT, ...) for learning and testing in Slicer. Because the datasets are hosted in a remote repository, a network connection is required to access them.
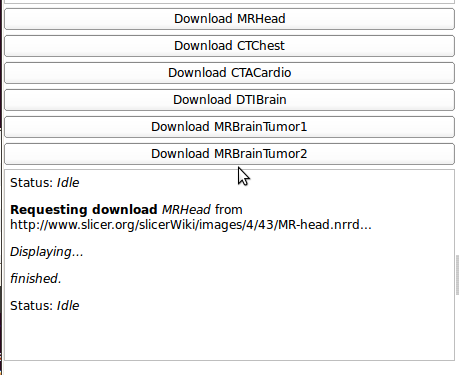
Use Cases
Examples:
- Use Case 1: Download an MR Volume to experimenting with the EMSegmenter.
- Use Case 2: Download a CT Volume for use with the Volume Rendering Module.
- Use Case 3: Download a DTI Volume for use with Tractography.
Tutorials
- Tutorial about loading and viewing data.
Panels and their use
This interface consists of a series of buttons for various download volumes. Status about downloads is displayed below the buttons.
Similar Modules
- The DICOM module can be used to access sample data on DICOM servers.
References
N/A
Information for Developers
This module is written in python, and relies on MRML's ability to access datasets via the http protocol.
The sample data is accessed from the wiki at this url: https://www.slicer.org/wiki/SampleData Modules can register their own sample data by following the example in the module.
| Section under construction. |


