Difference between revisions of "Documentation/4.1/Modules/FacetedVisualizer"
m |
(Prepend documentation/versioncheck template. See http://na-mic.org/Mantis/view.php?id=2887) |
||
| (15 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <noinclude>{{documentation/versioncheck}}</noinclude> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-header}} | {{documentation/{{documentation/version}}/module-header}} | ||
| Line 9: | Line 10: | ||
Extension: [[Documentation/{{documentation/version}}/Extensions/FacetedVisualizer|FacetedVisualizer]]<br> | Extension: [[Documentation/{{documentation/version}}/Extensions/FacetedVisualizer|FacetedVisualizer]]<br> | ||
Acknowledgments: | Acknowledgments: | ||
| − | This work is part of the Neuroimage Analysis Center (NAC), an NIBIB National Resource Center, Grant P41 EB015902.<br> | + | This work is part of the [http://nac.spl.harvard.edu Neuroimage Analysis Center (NAC)], an NIBIB National Resource Center, Grant P41 EB015902.<br> |
This work is based on the [http://sig.biostr.washington.edu/projects/fm/ Foundational Model of Anatomy (FMA) Version 3.0] from the Structural Informatics Group at the University of Washington.<br> | This work is based on the [http://sig.biostr.washington.edu/projects/fm/ Foundational Model of Anatomy (FMA) Version 3.0] from the Structural Informatics Group at the University of Washington.<br> | ||
Author: Harini Veeraraghavan ({{collaborator|name|ge}})<br> | Author: Harini Veeraraghavan ({{collaborator|name|ge}})<br> | ||
| − | Contributor1: Mike Halle ({{collaborator|name| | + | Contributor1: Mike Halle ({{collaborator|name|spl}})<br> |
Contributor2: Jim Miller ({{collaborator|name|ge}})<br> | Contributor2: Jim Miller ({{collaborator|name|ge}})<br> | ||
Contact: Jim Miller, <email>millerjv@ge.com</email><br> | Contact: Jim Miller, <email>millerjv@ge.com</email><br> | ||
| Line 18: | Line 19: | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
|{{collaborator|logo|ge}}|{{collaborator|longname|ge}} | |{{collaborator|logo|ge}}|{{collaborator|longname|ge}} | ||
| − | |{{collaborator|logo| | + | |{{collaborator|logo|spl}}|{{collaborator|longname|spl}} |
|{{collaborator|logo|nac}}|{{collaborator|longname|nac}} | |{{collaborator|logo|nac}}|{{collaborator|longname|nac}} | ||
}} | }} | ||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| + | Faceted Visualizer enables a user to create flexible visualizations using Slicer by combining a 3D digital atlas and 3D scene models with an ontology. This visualizer is primarily intended as a learning tool (e.g. neuroanatomy) and allows users to employ "drill-down" search strategies starting from a specific user typed query by showing the related options extracted by searching the ontology. Currently, the faceted visualizer has been tested with the [http://www.spl.harvard.edu/publications/item/view/2037 SPL-PNL Brain atlas] and the [http://www.spl.harvard.edu/publications/item/view/1918 SPL Abdominal atlas] and the [http://sig.biostr.washington.edu/projects/fm/ FMA ontology]. The Radlex ontology is currently not supported by the visualizer. | ||
| + | |||
| + | The Faceted Visualizer uses a derivative of the FMA, limited to a ''subset'' of concepts and terms that are needed to support a search based paradigm to control the visualization of segmented and labeled anatomy. The information extracted from the FMA has been augmented with select "functional system" terms. The ontology is represented as a triple store <Subject, Predicate, Object>. The ontology for the Faceted Visualizer can be downloaded [http://slicer.kitware.com/midas3/item/3094 here]. | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| + | The faceted visualizer integrates an ontology with a 3D Atlas for combined visualization with relevant textual information extracted from the ontology. The visualizer module employs a hierarchical search on the ontology to obtain the visualizations. The visualizer supports simple (e.g. "putamen", "motor system", etc), to complex queries ("liver;arterial supply + stomach + kidney;arterial supply"). | ||
| Line 31: | Line 36: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
| − | {| | + | {|align="center" |
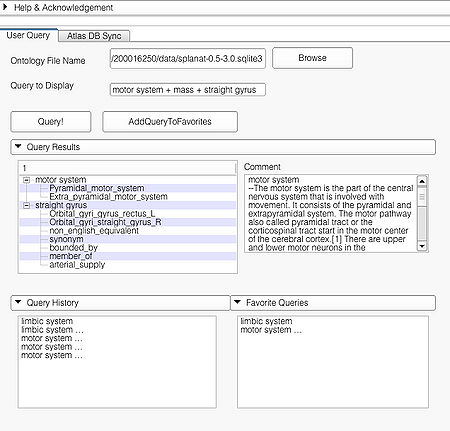
| − | |[[ | + | |[[image:ComplexQueryAPI.jpg|thumb|450px| Query page and results of a complex query "motor system + mass + straight gyrus"]] |
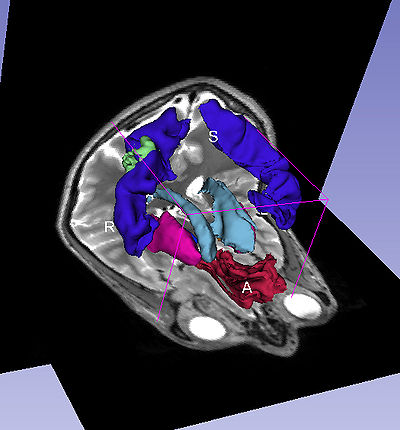
| − | |[[ | + | |[[image:ComplexQueryWithImage.jpg|thumb|400px| Visualization of complex query "motor system + mass + straight gyrus"]] |
| − | |||
|} | |} | ||
| Line 42: | Line 46: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
| + | The GUI consists of two main panels. The "User Query" page and "AtlasDBSync" page. The "User Query page" is where the user interacts with the visualizer to produce the visualizations. The "AtlasDBSync" page helps to integrate the ontology with the 3D models contained in the atlas. The visualizer module automatically tries to find appropriate mappings of the 3D models with the ontology. But for models where exact matches aren't found, the visualizer module tries to find all the matching entries from the DB. The interface in the "AtlasDBSync" page allows user to check the mappings found by the module and set up the correct mappings between the ontology and the DB. | ||
| − | {| | + | {{documentation/{{documentation/version}}/module-subsection|User Query Page}} |
| − | | | + | {|align="right" |
| − | |[[ | + | |[[image:APIUserQueryPageGUI.jpg|thumb|500px|User Page GUI]] |
| + | |} | ||
| + | |||
| + | This page is useful for creating visualizations based on user typed queries. The user inputs consist of the '''Ontology File Name''' and '''Query To Display'''. The button '''Query''' fires a query and causes the module to start processing the user query for visualization. The button '''AddQueryToFavorites''' adds the last typed user query to the list of favorite queries. The favorite query list is shown in the bottom panel '''Favorite Queries'''. Additionally, the interface also shows a history of typed queries in the panel '''Query History'''. | ||
| + | |||
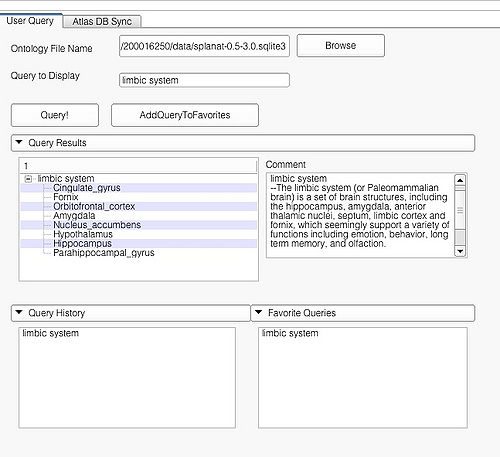
| + | The '''Query Results''' panel shows the result of a query, consisting of all the associated predicates and parts for the query. The Figure ''User Page GUI'' shows the result of a simple query "''limbic system''".Additionally, the interface also shows a '''Comment''' that contains any relevant textual information about the query obtained from the ontology. The user can refine a search by selecting from the query result. Optionally, the user can also select by clicking on any selection from the '''Query History''' or '''Favorite Queries''' panel to redo search and visualization. The interface supports the multiple selections to form complex queries. The visualization resulting by searching with the query "Limbic system" is shown below. | ||
| + | {|align="center" | ||
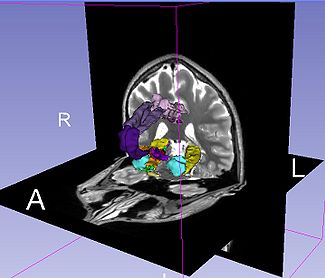
| + | |[[image:LimbicSystemWithImage.jpg|thumb|325px| Visualization of Query "limbic system"]] | ||
| + | |} | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsubsection|Querying Specifics}} | ||
| + | The faceted visualization module supports the following types of queries: | ||
| + | * Simple Queries e.g. "putamen", "limbic system" | ||
| + | * Complex Queries e.g. "limbic system + mass", and | ||
| + | * Specialized Queries e.g. "liver;arterial_supply" | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsubsubsection|Simple Queries}} | ||
| + | Simple query is a self-contained individual string that is matched in entirely with the ontology. An example of a simple query is "limbic system". The result of this query is shown above. | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsubsubsection|Complex Queries}} | ||
| + | A complex query contains two or more simple or specialized queries. A complex query is typically specified by using a '''"+"'''. Note that the '''"+"''' does not signify an ANDing of the search terms. Instead each string separated by the '''"+"''' is searched as an individual query in the ontology. The result of using a complex query is shown below. The Application interface shows the results for all the terms that are matched in the ontology (i.e. the motor system and straight gyrus.) Unmatched terms such as the user added models (e.g. mass) are only displayed. The figure shows an example of a complex query comprised of simple queries. | ||
| + | {|align="center" | ||
| + | |[[image:ComplexQueryAPI.jpg|thumb|450px| Query page and results of a complex query "motor system + mass + straight gyrus"]] | ||
| + | |[[image:ComplexQueryWithImage.jpg|thumb|400px| Visualization of complex query "motor system + mass + straight gyrus"]] | ||
| + | |} | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsubsubsection|Specialized Queries}} | ||
| + | A specialized query helps to search for a query with additional constraints typically using an ANDing operation. Specialized query is specified using a '''";"''' between the search terms. In terms of search in the ontology, the '''";"''' will either result in a search involving the first term as the '''Subject''' or '''Object''' and the second term as the '''Predicate'''(e.g. "straight gyrus;synonym"). In the case where the second term is not a predicate but a part of the preceding term, e.g. "limbic system;Fornix", the visualizer logic automatically detects that the succeeding term is an individual search term and produces the visualization using the succeeding term as the search string resulting in the visualization of query "Fornix". | ||
| + | |||
| + | Specialized queries can be combined with other simple or specialized queries to form complex queries (e.g. "motor system + straight gyrus;synonym"). An example of a complex query using specialized and simple query terms is shown below. | ||
| + | |||
| + | {|align="center" | ||
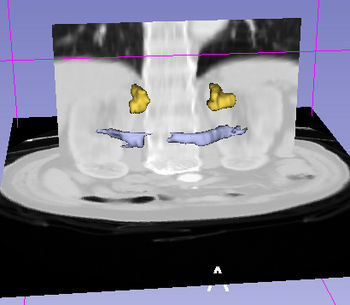
| + | |[[image:CompleQueryAbdominalRes.jpg|thumb|350px|Complex query result using simple and specialized query terms "spleen + pancreas;arterial_supply"]] | ||
| + | |[[image:ComplexQueryAbdominalWithImage.jpg|thumb|350px | Visualization of complex query containing simple and specialized terms "spleen + pancreas;arterial_supply"]] | ||
| + | |} | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsubsection|Query Interface}} | ||
| + | The user can query by typing the query in the Query to Display box or by selecting from one of '''Query Results''', '''Query History''', and '''Favorite Query''' panels. | ||
| + | All the panels support multiple selection. Multiple selection will result in a complex query. An example of multiple selection from the '''Query Results''' panels is shown below. | ||
| + | However, it is not possible to create a complex query by selecting from multiple panels (e.g. selecting "some string in Query Results" and "some other string in Query History"). | ||
| + | |||
| + | |||
| + | Selecting a string from the '''Query Result''' panel will result in creating a specialized query. Example of such a selection is shown below. Multiple selections from the '''"Query Result"''' pane will result in a complex query using specialized query terms as shown. | ||
| + | {|align="center" | ||
| + | |[[image:ComplexQueryAbdominal.jpg|thumb|350px|Complex query creation from the Query Results pane]] | ||
| + | |} | ||
| + | |||
| + | Queries can also be obtained by selecting from the '''Query History''' pane and the '''Favorite Queries''' pane. Both support multiple selections as shown below. | ||
| + | {|align="center" | ||
| + | |[[image:QueryInterfaceHistoryPane.jpg|thumb|350px|Complex query through multiple selection from Query Histroy]] | ||
| + | |[[image:QueryInterfaceFavorites.jpg|thumb|350px|Complex query through multiple selection from Favorite Queries]] | ||
| + | |} | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsection|AtlasDBSync Page}} | ||
| + | The faceted visualizer module does not require the user to explicitly set the mappings between the the 3D scene atlas and the ontology database before use. Everytime a scene is loaded and an ontology file is set, the module automatically tries to compute matches between the models in the atlas with terms in the ontology. Matching is done in two-passes. In the first pass, the model names (excluding numbers etc) in the atlas are matched exactly (except for white spaces and case) with the terms in the ontology. When a match is found, the mapping between a given model (name) and the database term is stored. For those models where no match is found, a second-pass match is performed. In the second-pass, a relaxed matching is performed where substrings of the model name string is matched with the terms in the ontology. This step usually creates non-unique matches. As a result, the mappings are not set automatically. Instead the matching options from the ontology are stored and can be selected by the user. The user can accomplish this from the '''"AtlasDBSync"''' page. | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsubsection|Application interface}} | ||
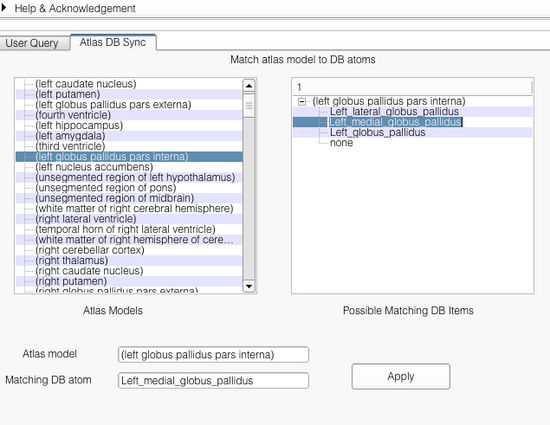
| + | The application interface is shown below. As shown, the interface shows the list of all the models found in the atlas. Selecting an atlas model on the left pane shows the corresponding matches found in the right pane. The user can set the match by selecting appropriate matching string from the right pane. Upon selection from the right pane, both the atlas model and the selected ontology term are shown in the bottom '''Atlas model''' and '''Matching DB atom'''. The user can confirm the match by clicking on the button '''Apply'''. | ||
| + | {|align="center" | ||
| + | |[[image:AtlasDBSyncGUI.jpg|thumb|550px|Synchronize Atlas With Ontology Page]] | ||
|} | |} | ||
| Line 53: | Line 117: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | |||
| − | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
| + | <!--{{documentation/{{documentation/version}}/module-developerinfo}}--> | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsection|Algorithm Details}} | ||
| + | The two main components of the faceted visualization module are: | ||
| + | * Synchronization of the 3D atlas with the ontology and | ||
| + | * Hierarchical-search based query visualization. | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsubsection|Background: Ontology}} | ||
| + | The ontology is essentially a SQL database where the various terms are organized as triples '''<Subject, Predicate, Object>'''. In this work, we employ the FMA ontology for searching and visualization. As the only elements present in the ontology are the atoms and their relations expressed as triples, we employ hierarchical search through the terms to extract taxonomies and functional organizations of the searched queries. | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsubsection|Synchronization of the 3D atlas with ontology}} | ||
| + | This is the entry point in the algorithm. As a first step, the synchronizer tries to match all the models in the 3D atlas and the models added by the user in the scene with the ontology. In this work, we assume that the model names used for the various atlases will be names corresponding to the organ or neuroanatomy part being displayed. Hence, for synchronization, we try to match the model names with the atoms in the ontology. We employ a two-step approach for matching. In the first step, the model names are matched exactly with the atoms in the ontology with the exception of white space and character case (upper or lower) differences. To match, the synchronizer searches in both the subject and object location of the various terms. | ||
| + | Exact matching ensures that we get the best possible matches. | ||
| + | |||
| + | However, there can still be some differences in the naming convention used in the models and the ontology resulting in an absence of match. Examples include '''"pineal body"''' used in the SPL_PNL Brain atlas and '''"Pineal_gland"''' used in the FMA ontology, '''"left globus pallidus pars interna"''', and '''"Left_medial_globus_pallidus"''', used in the SPL_PNL atlas and the FMA ontology, respectively. To deal with the different naming of the models than in the ontology, we employ a second step searching which employs a relaxed matching. The relaxed match tries to match the individual components of the string with the database. For example, the string '''pineal body''' will result in two different SQL queries: (i) ''SELECT * from resources where subject like '%Pineal%' or object like '%Pineal%'', and (ii) ''SELECT * from resources where subject like '%body%' or object like '%body%''. The relaxed search results in none, one, or many matches. All the matching atoms are stored and then displayed to the user for selection using the Atlas Database Synchronization Interface in the GUI. Optionally, the user can also check the accuracy of matches for the matched models and database atoms and correct them if necessary. | ||
| + | |||
| + | The synchronization step is performed only once during the entry to the program or whenever the user modifies the database file to be used by the module. As a result of the synchronization, a mapping between the 3D atlas models and their corresponding names occurring in the ontology is stored. Every time the user changes the database file that will be used, the previous mappings are removed to create a new one. | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsubsection|Hierarchical-search based query visualization}} | ||
| + | This is the second step for visualization and is performed every time the user enters a new query. By hierarchical search we mean a recursive search through the ontology using the results of each query. The hierarchical (or in other words recursive) search is performed to extract all the relevant terms relating to a search query. This is particularly important when the query involves the search for a structure that is composed of various parts, e.g. thalamus, diencephalon, motor system, GI, etc. | ||
| + | |||
| + | As the user queries can be composed of simple or complex queries, first the query string is parsed to extract sub-queries that may be simple or specialized queries. Next, using each sub-query, a SQL query is formed to search the database. As the searched query string can occur in either the '''Subject''' or '''Object''' part in the database, the query is formulated to capture both cases. Next, the result of the query is parsed to extract the relevant atoms to be reprocessed for querying. | ||
| + | |||
| + | When the query is a simple query, the resulting SQL query is of the form ''SELECT * from resources where Subject = 'query' or Object = 'query''. When the query is a specialized query, the resulting SQL query is of the form ''SELECT * from resources where Subject = 'firstPartOfQuery' and Predicate = 'secondPartOfQuery' or 'Object = 'firstPartOfQuery' and Predicate = 'secondPartOfQuery'''. Subsequent recursions will employ the simple query form. | ||
| + | |||
| + | Since the goal of the search is only to extract all the relevant parts of a given structure, the recursive search is repeated only for the atoms that are contained in ''relations'' that signify a part relationship. Examples of such relationships occurring in the FMA ontology include: ''constitutional_part'', ''regional_part'', ''systemic_part'', ''member'', and ''subClass'', etc when the query string occurred in the '''Subject''' portion of the query result. On the other hand, when the query string occurred in the '''Object''' portion of the query result, corresponding relations such as ''constitutional_part_of'', ''regional_part_of' etc are used. In the former case, the atoms occurring in the '''Object''' portion of the search result relations are used for re-querying, while in the latter case, the atoms occurring in the '''Subject''' portion of search result relations are used. Subsequent searches will involve only searches where the new query resulting from the result atoms occurs in the '''Subject''' portion of the database relations. This ensures to avoid looping of the searches. The recursive search stops when there are no more ''relation'' terms occurring in the query result. | ||
| + | |||
| + | Each successive SQL query results in one or more relations. The appropriate objects or subjects from those relations (based on the occurrence of the query string) are then compared with the models-database relation to extract the relevant models in the atlas for displaying. Additionally, all the predicates that are relevant to the user query and the recovered parts are stored for displaying textual information and allowing drill-down querying from the GUI. | ||
| + | |||
| + | {{documentation/{{documentation/version}}/module-subsection|Limitations and Future Work}} | ||
| + | * Current implementation does not support any partial matching of the query and the ontology search result with the Atlas models. For ex. query "stomach" will return an empty visualization using the abdominal atlas as the model name in the atlas is "Model_13_stomach_and_duodenum". The query term needs to be "stomach and duodenum" to return a visualization. | ||
| + | * Current implementation does not allow the currently found atlas and database bindings to be stored either in the ontology or in the atlas. This can be cumbersome for the user as they may have to redo some of the mappings every time the visualizer is loaded with a specific atlas and an ontology. This was done deliberately to prevent cluttering the database with every new atlas. On the other hand, in the future, we may store the bindings as attributes in the MRML Scene for each specific ontology. | ||
| + | * Current implementation will not detect any changes to the scene that are made after initializing the faceted visualizer with a database file. Any new imports or additions, deletions of models will be ignored by the module. In the future, we may try to automatically detect changes to the scene and modify the mappings in run-time. | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 63: | Line 160: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-footer}} | {{documentation/{{documentation/version}}/module-footer}} | ||
| − | [[Category:Documentation/{{documentation/version}}/Modules/ | + | [[Category:Documentation/{{documentation/version}}/Modules/Informatics]] |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 07:24, 14 June 2013
Home < Documentation < 4.1 < Modules < FacetedVisualizer
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: FacetedVisualizer | |||||||
|
Faceted Visualizer enables a user to create flexible visualizations using Slicer by combining a 3D digital atlas and 3D scene models with an ontology. This visualizer is primarily intended as a learning tool (e.g. neuroanatomy) and allows users to employ "drill-down" search strategies starting from a specific user typed query by showing the related options extracted by searching the ontology. Currently, the faceted visualizer has been tested with the SPL-PNL Brain atlas and the SPL Abdominal atlas and the FMA ontology. The Radlex ontology is currently not supported by the visualizer.
The Faceted Visualizer uses a derivative of the FMA, limited to a subset of concepts and terms that are needed to support a search based paradigm to control the visualization of segmented and labeled anatomy. The information extracted from the FMA has been augmented with select "functional system" terms. The ontology is represented as a triple store <Subject, Predicate, Object>. The ontology for the Faceted Visualizer can be downloaded here.
Module Description
The faceted visualizer integrates an ontology with a 3D Atlas for combined visualization with relevant textual information extracted from the ontology. The visualizer module employs a hierarchical search on the ontology to obtain the visualizations. The visualizer supports simple (e.g. "putamen", "motor system", etc), to complex queries ("liver;arterial supply + stomach + kidney;arterial supply").
Use Cases
Tutorials
Panels and their use
The GUI consists of two main panels. The "User Query" page and "AtlasDBSync" page. The "User Query page" is where the user interacts with the visualizer to produce the visualizations. The "AtlasDBSync" page helps to integrate the ontology with the 3D models contained in the atlas. The visualizer module automatically tries to find appropriate mappings of the 3D models with the ontology. But for models where exact matches aren't found, the visualizer module tries to find all the matching entries from the DB. The interface in the "AtlasDBSync" page allows user to check the mappings found by the module and set up the correct mappings between the ontology and the DB.
User Query Page
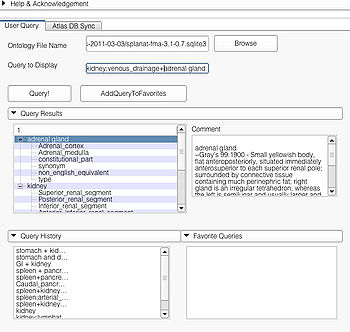
This page is useful for creating visualizations based on user typed queries. The user inputs consist of the Ontology File Name and Query To Display. The button Query fires a query and causes the module to start processing the user query for visualization. The button AddQueryToFavorites adds the last typed user query to the list of favorite queries. The favorite query list is shown in the bottom panel Favorite Queries. Additionally, the interface also shows a history of typed queries in the panel Query History.
The Query Results panel shows the result of a query, consisting of all the associated predicates and parts for the query. The Figure User Page GUI shows the result of a simple query "limbic system".Additionally, the interface also shows a Comment that contains any relevant textual information about the query obtained from the ontology. The user can refine a search by selecting from the query result. Optionally, the user can also select by clicking on any selection from the Query History or Favorite Queries panel to redo search and visualization. The interface supports the multiple selections to form complex queries. The visualization resulting by searching with the query "Limbic system" is shown below.
Querying Specifics
The faceted visualization module supports the following types of queries:
- Simple Queries e.g. "putamen", "limbic system"
- Complex Queries e.g. "limbic system + mass", and
- Specialized Queries e.g. "liver;arterial_supply"
Simple Queries
Simple query is a self-contained individual string that is matched in entirely with the ontology. An example of a simple query is "limbic system". The result of this query is shown above.
Complex Queries
A complex query contains two or more simple or specialized queries. A complex query is typically specified by using a "+". Note that the "+" does not signify an ANDing of the search terms. Instead each string separated by the "+" is searched as an individual query in the ontology. The result of using a complex query is shown below. The Application interface shows the results for all the terms that are matched in the ontology (i.e. the motor system and straight gyrus.) Unmatched terms such as the user added models (e.g. mass) are only displayed. The figure shows an example of a complex query comprised of simple queries.
Specialized Queries
A specialized query helps to search for a query with additional constraints typically using an ANDing operation. Specialized query is specified using a ";" between the search terms. In terms of search in the ontology, the ";" will either result in a search involving the first term as the Subject or Object and the second term as the Predicate(e.g. "straight gyrus;synonym"). In the case where the second term is not a predicate but a part of the preceding term, e.g. "limbic system;Fornix", the visualizer logic automatically detects that the succeeding term is an individual search term and produces the visualization using the succeeding term as the search string resulting in the visualization of query "Fornix".
Specialized queries can be combined with other simple or specialized queries to form complex queries (e.g. "motor system + straight gyrus;synonym"). An example of a complex query using specialized and simple query terms is shown below.
Query Interface
The user can query by typing the query in the Query to Display box or by selecting from one of Query Results, Query History, and Favorite Query panels. All the panels support multiple selection. Multiple selection will result in a complex query. An example of multiple selection from the Query Results panels is shown below. However, it is not possible to create a complex query by selecting from multiple panels (e.g. selecting "some string in Query Results" and "some other string in Query History").
Selecting a string from the Query Result panel will result in creating a specialized query. Example of such a selection is shown below. Multiple selections from the "Query Result" pane will result in a complex query using specialized query terms as shown.
Queries can also be obtained by selecting from the Query History pane and the Favorite Queries pane. Both support multiple selections as shown below.
AtlasDBSync Page
The faceted visualizer module does not require the user to explicitly set the mappings between the the 3D scene atlas and the ontology database before use. Everytime a scene is loaded and an ontology file is set, the module automatically tries to compute matches between the models in the atlas with terms in the ontology. Matching is done in two-passes. In the first pass, the model names (excluding numbers etc) in the atlas are matched exactly (except for white spaces and case) with the terms in the ontology. When a match is found, the mapping between a given model (name) and the database term is stored. For those models where no match is found, a second-pass match is performed. In the second-pass, a relaxed matching is performed where substrings of the model name string is matched with the terms in the ontology. This step usually creates non-unique matches. As a result, the mappings are not set automatically. Instead the matching options from the ontology are stored and can be selected by the user. The user can accomplish this from the "AtlasDBSync" page.
Application interface
The application interface is shown below. As shown, the interface shows the list of all the models found in the atlas. Selecting an atlas model on the left pane shows the corresponding matches found in the right pane. The user can set the match by selecting appropriate matching string from the right pane. Upon selection from the right pane, both the atlas model and the selected ontology term are shown in the bottom Atlas model and Matching DB atom. The user can confirm the match by clicking on the button Apply.
Similar Modules
References
Information for Developers
Algorithm Details
The two main components of the faceted visualization module are:
- Synchronization of the 3D atlas with the ontology and
- Hierarchical-search based query visualization.
Background: Ontology
The ontology is essentially a SQL database where the various terms are organized as triples <Subject, Predicate, Object>. In this work, we employ the FMA ontology for searching and visualization. As the only elements present in the ontology are the atoms and their relations expressed as triples, we employ hierarchical search through the terms to extract taxonomies and functional organizations of the searched queries.
Synchronization of the 3D atlas with ontology
This is the entry point in the algorithm. As a first step, the synchronizer tries to match all the models in the 3D atlas and the models added by the user in the scene with the ontology. In this work, we assume that the model names used for the various atlases will be names corresponding to the organ or neuroanatomy part being displayed. Hence, for synchronization, we try to match the model names with the atoms in the ontology. We employ a two-step approach for matching. In the first step, the model names are matched exactly with the atoms in the ontology with the exception of white space and character case (upper or lower) differences. To match, the synchronizer searches in both the subject and object location of the various terms. Exact matching ensures that we get the best possible matches.
However, there can still be some differences in the naming convention used in the models and the ontology resulting in an absence of match. Examples include "pineal body" used in the SPL_PNL Brain atlas and "Pineal_gland" used in the FMA ontology, "left globus pallidus pars interna", and "Left_medial_globus_pallidus", used in the SPL_PNL atlas and the FMA ontology, respectively. To deal with the different naming of the models than in the ontology, we employ a second step searching which employs a relaxed matching. The relaxed match tries to match the individual components of the string with the database. For example, the string pineal body will result in two different SQL queries: (i) SELECT * from resources where subject like '%Pineal%' or object like '%Pineal%, and (ii) SELECT * from resources where subject like '%body%' or object like '%body%. The relaxed search results in none, one, or many matches. All the matching atoms are stored and then displayed to the user for selection using the Atlas Database Synchronization Interface in the GUI. Optionally, the user can also check the accuracy of matches for the matched models and database atoms and correct them if necessary.
The synchronization step is performed only once during the entry to the program or whenever the user modifies the database file to be used by the module. As a result of the synchronization, a mapping between the 3D atlas models and their corresponding names occurring in the ontology is stored. Every time the user changes the database file that will be used, the previous mappings are removed to create a new one.
Hierarchical-search based query visualization
This is the second step for visualization and is performed every time the user enters a new query. By hierarchical search we mean a recursive search through the ontology using the results of each query. The hierarchical (or in other words recursive) search is performed to extract all the relevant terms relating to a search query. This is particularly important when the query involves the search for a structure that is composed of various parts, e.g. thalamus, diencephalon, motor system, GI, etc.
As the user queries can be composed of simple or complex queries, first the query string is parsed to extract sub-queries that may be simple or specialized queries. Next, using each sub-query, a SQL query is formed to search the database. As the searched query string can occur in either the Subject or Object part in the database, the query is formulated to capture both cases. Next, the result of the query is parsed to extract the relevant atoms to be reprocessed for querying.
When the query is a simple query, the resulting SQL query is of the form SELECT * from resources where Subject = 'query' or Object = 'query. When the query is a specialized query, the resulting SQL query is of the form SELECT * from resources where Subject = 'firstPartOfQuery' and Predicate = 'secondPartOfQuery' or 'Object = 'firstPartOfQuery' and Predicate = 'secondPartOfQuery'. Subsequent recursions will employ the simple query form.
Since the goal of the search is only to extract all the relevant parts of a given structure, the recursive search is repeated only for the atoms that are contained in relations that signify a part relationship. Examples of such relationships occurring in the FMA ontology include: constitutional_part, regional_part, systemic_part, member, and subClass, etc when the query string occurred in the Subject portion of the query result. On the other hand, when the query string occurred in the Object portion of the query result, corresponding relations such as constitutional_part_of, regional_part_of' etc are used. In the former case, the atoms occurring in the Object portion of the search result relations are used for re-querying, while in the latter case, the atoms occurring in the Subject portion of search result relations are used. Subsequent searches will involve only searches where the new query resulting from the result atoms occurs in the Subject portion of the database relations. This ensures to avoid looping of the searches. The recursive search stops when there are no more relation terms occurring in the query result.
Each successive SQL query results in one or more relations. The appropriate objects or subjects from those relations (based on the occurrence of the query string) are then compared with the models-database relation to extract the relevant models in the atlas for displaying. Additionally, all the predicates that are relevant to the user query and the recovered parts are stored for displaying textual information and allowing drill-down querying from the GUI.
Limitations and Future Work
- Current implementation does not support any partial matching of the query and the ontology search result with the Atlas models. For ex. query "stomach" will return an empty visualization using the abdominal atlas as the model name in the atlas is "Model_13_stomach_and_duodenum". The query term needs to be "stomach and duodenum" to return a visualization.
- Current implementation does not allow the currently found atlas and database bindings to be stored either in the ontology or in the atlas. This can be cumbersome for the user as they may have to redo some of the mappings every time the visualizer is loaded with a specific atlas and an ontology. This was done deliberately to prevent cluttering the database with every new atlas. On the other hand, in the future, we may store the bindings as attributes in the MRML Scene for each specific ontology.
- Current implementation will not detect any changes to the scene that are made after initializing the faceted visualizer with a database file. Any new imports or additions, deletions of models will be ignored by the module. In the future, we may try to automatically detect changes to the scene and modify the mappings in run-time.
Information for Developers
| Section under construction. |