Difference between revisions of "Documentation/4.0/Training"
From Slicer Wiki
| (5 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <noinclude>{{documentation/versioncheck}}</noinclude> | ||
__TOC__ | __TOC__ | ||
| Line 9: | Line 10: | ||
*For questions related to the Slicer4 Compendium, please send an e-mail to '''[http://www.na-mic.org/Wiki/index.php/User:SPujol Sonia Pujol, Ph.D]''' | *For questions related to the Slicer4 Compendium, please send an e-mail to '''[http://www.na-mic.org/Wiki/index.php/User:SPujol Sonia Pujol, Ph.D]''' | ||
| + | For information on 3D Slicer version 3.6, see [[Slicer_3.6:Training| the 3.6 Training Pages]]. | ||
=General Introduction= | =General Introduction= | ||
| Line 52: | Line 54: | ||
*Author: Sonia Pujol, Ph.D., Steve Pieper, Ph.D. | *Author: Sonia Pujol, Ph.D., Steve Pieper, Ph.D. | ||
*Audience: Developers | *Audience: Developers | ||
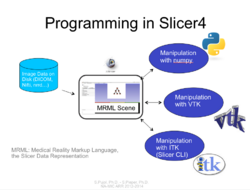
| − | *The [[Media:HelloPythonSlicer4.zip| HelloPython]] | + | *The [[Media:HelloPythonSlicer4.zip| HelloPython dataset]] contains three Python files and an MR scan of the brain. |
|align="right"| | |align="right"| | ||
[[Image:HelloPythonTutorial.png|right|250px|]] | [[Image:HelloPythonTutorial.png|right|250px|]] | ||
| Line 73: | Line 75: | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
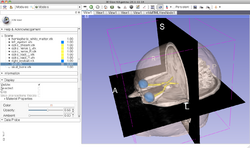
| − | *The [[Media:NeurosurgicalPlanning_SoniaPujol.pdf | Neurosurgical Planning tutorial]] course guides through | + | *The [[Media:NeurosurgicalPlanning_SoniaPujol.pdf | Neurosurgical Planning tutorial]] course guides through the generation of fiber tracts in the vicinity of a tumor. |
*Author: Sonia Pujol, Ph.D., Ron Kikinis, M.D. | *Author: Sonia Pujol, Ph.D., Ron Kikinis, M.D. | ||
*Audience: End-users and developers | *Audience: End-users and developers | ||
| Line 98: | Line 100: | ||
| | | | ||
*The [[media:Slicer4QuantitativeImaging.pdf| Slicer4 Quantitative Imaging tutorial]] guides through the use for Slicer for quantifying small volumetric changes in slow-growing tumors, and for calculating Standardized Uptake Value (SUV) from PET/CT data. | *The [[media:Slicer4QuantitativeImaging.pdf| Slicer4 Quantitative Imaging tutorial]] guides through the use for Slicer for quantifying small volumetric changes in slow-growing tumors, and for calculating Standardized Uptake Value (SUV) from PET/CT data. | ||
| − | *Authors: Jeffrey Yap, Ph.D., Ron Kikinis, M.D., Randy Gollub, M.D., Ph.D., Wendy Plesniak, Ph.D., Nicole Aucoin, B.Sc., Sonia Pujol, Ph.D., Valerie Humblet, Ph.D., Andriy Fedorov, Ph.D., Kilian | + | *Authors: Jeffrey Yap, Ph.D., Ron Kikinis, M.D., Randy Gollub, M.D., Ph.D., Wendy Plesniak, Ph.D., Nicole Aucoin, B.Sc., Sonia Pujol, Ph.D., Valerie Humblet, Ph.D., Andriy Fedorov, Ph.D., Kilian Pohl, Ph.D., Ender Konugolu, Ph.D. |
*Audience: Radiologists and users of Slicer who need a more comprehensive overview over Slicer4 quantitative imaging capabilities. | *Audience: Radiologists and users of Slicer who need a more comprehensive overview over Slicer4 quantitative imaging capabilities. | ||
*The [[media:PETCTFusion-TutorialData.zip| PETCTFusion]] and [[media:ChangeTracker2011.zip|Change Tracker]] datasets contain a series of MR, CT and PET data. | *The [[media:PETCTFusion-TutorialData.zip| PETCTFusion]] and [[media:ChangeTracker2011.zip|Change Tracker]] datasets contain a series of MR, CT and PET data. | ||
| Line 104: | Line 106: | ||
[[Image:Slicer4_QuantitativeImaging.png|right|250px|]] | [[Image:Slicer4_QuantitativeImaging.png|right|250px|]] | ||
|} | |} | ||
| − | |||
=Additional resources = | =Additional resources = | ||
{|width="100%" | {|width="100%" | ||
| | | | ||
| − | *This Intro to Slicer4.0 webinar provides an introduction to 3DSlicer, and demonstrates core functionalities such as loading, visualizing and saving data. Basic processing tools, including manual registration, manual segmentation and tractography tools are also highlighted. This | + | *This Intro to Slicer4.0 [http://vimeo.com/37671358 webinar] provides an introduction to 3DSlicer, and demonstrates core functionalities such as loading, visualizing and saving data. Basic processing tools, including manual registration, manual segmentation and tractography tools are also highlighted. This webinar is a general overview. For in depth information see the modules above and the documentation pages. |
*Authors: Julien Finet, M.S., Steve Pieper, Ph.D., Jean-Christophe Fillion-Robin, M.S. | *Authors: Julien Finet, M.S., Steve Pieper, Ph.D., Jean-Christophe Fillion-Robin, M.S. | ||
*Audience: First time users interested in a broad overview of Slicer’s features and tools. | *Audience: First time users interested in a broad overview of Slicer’s features and tools. | ||
|align="right"| | |align="right"| | ||
| − | [[Image: | + | [[Image:Webinar.png|250px]] |
|} | |} | ||
Latest revision as of 19:30, 25 April 2014
Home < Documentation < 4.0 < Training
|
For the latest Slicer documentation, visit the read-the-docs. |
Contents
Introduction: Slicer 4.0 Tutorials
- This page contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version 4.0 release) to accomplish certain tasks.
- For tutorials for other versions of Slicer, please visit the Slicer training portal.
- For "reference manual" style documentation, please visit the Slicer 4.0 documentation page
- For questions related to the Slicer4 Compendium, please send an e-mail to Sonia Pujol, Ph.D
For information on 3D Slicer version 3.6, see the 3.6 Training Pages.
General Introduction
Slicer Welcome Tutorial
|
Slicer4Minute Tutorial
|
Slicer4 Data Loading and 3D Visualization
|
Tutorials for software developers
Slicer4 Programming Tutorial
|
Specific functions
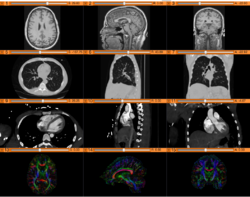
Slicer4 Diffusion Tensor Imaging Tutorial
|
Slicer4 Neurosurgical Planning Tutorial
|
Slicer4 3D Visualization of DICOM images for Radiology Applications
|
Slicer4 Quantitative Imaging tutorial
|
Additional resources
|