Difference between revisions of "Slicer 3.6:Training"
From Slicer Wiki
m Tag: 2017 source edit |
|||
| (119 intermediate revisions by 11 users not shown) | |||
| Line 1: | Line 1: | ||
=Slicer 3.6 Tutorials= | =Slicer 3.6 Tutorials= | ||
| − | * The following table contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version 3.6 release) to accomplish certain tasks. | + | *The following table contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version 3.6 release) to accomplish certain tasks. |
| − | * For tutorials for other versions of Slicer, please visit the [[Training| Slicer training portal]]. | + | *For tutorials for other versions of Slicer, please visit the [[Training| Slicer training portal]]. |
| − | *For questions related to the Slicer3 Compendium, please send an e-mail to [http://www.na-mic.org/Wiki/index.php/User:SPujol Sonia Pujol, Ph.D] | + | *For questions related to the Slicer3 Compendium, please send an e-mail to '''[http://www.na-mic.org/Wiki/index.php/User:SPujol Sonia Pujol, Ph.D]''' |
| + | <br> | ||
| − | {| border="1" cellpadding=" | + | {| border="1" cellpadding="3" width="1000px" |
|- style="background:#CCFF99; color:black; font-size:130%" align="center" | |- style="background:#CCFF99; color:black; font-size:130%" align="center" | ||
| − | |'''Category''' | + | | style="width:100px" |'''Category''' |
| − | |'''Tutorial''' | + | | style="width:350px" |'''Tutorial''' |
| − | |'''Sample Data''' | + | | style="width:350px" |'''Sample Data''' |
| − | |'''Image''' | + | | style="width:250px" |'''Image''' |
|- | |- | ||
| − | | style="background:#FFFFCC; color:blue; font-size:110%" align="center"| | + | | id="Slicer3MinuteTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Core''' |
| − | | style="background:#FFFFCC; color:black" valign="top"| '''[[ | + | | style="background:#FFFFCC; color:black" valign="top" |'''[[Media:Slicer3Minute SoniaPujol 3.6.1.pdf| Slicer3Minute Tutorial]]'''<br><br><br>The Slicer3Minute tutorial is an introduction to the advanced 3D visualization capabilities of Slicer3.6. <br><br>'''Audience:''' First time users. |
| − | | style="background:#FFFFCC; color:black" valign="top" | '''[http://www.slicer.org/ | + | | style="background:#FFFFCC; color:black" valign="top" |'''[http://www.slicer.org/w/img_auth.php/5/51/Slicer3MinuteDataset.zip Slicer3Minute Data]'''<br><br> The Slicer3Minute dataset contains an MR scan of the brain and 3D reconstructions of the anatomy |
| − | | style="background:#FFFFCC; color:black" align="center"| [[Image:Slicer3Minute_3.6RC2.png | | + | | style="background:#FFFFCC; color:black" align="center" |[[Image:Slicer3Minute_3.6RC2.png |250px]] |
|- | |- | ||
| − | | style="background:#FFFFCC; color:blue; font-size:110%" align="center"| | + | | id="Slicer3VisualizationTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Core''' |
| − | | style="background:#FFFFCC; color:black" valign="top"| '''[[ | + | | style="background:#FFFFCC; color:black" valign="top" |'''[[Media:Slicer3 DataLoadingAndVisualization UCSF2010 SoniaPujol.pdf| Slicer3Visualization Tutorial]]'''<br><br><br>The Slicer3Visualization tutorial guides through 3D data loading and visualization in Slicer3.6.<br><br>'''Audience:''' All beginners including clinicians, scientists, engineers and programmers. |
| − | | style="background:#FFFFCC; color:black" valign="top" | '''[http://www.slicer.org/ | + | | style="background:#FFFFCC; color:black" valign="top" |'''[http://www.slicer.org/w/img_auth.php/6/61/Slicer3VisualizationDataset.zip Slicer3Visualization Data]'''<br><br>The Slicer3VisualizationDataset contains two MR scans of the brain, a pre-computed labelmap and 3D reconstructions of the anatomy. |
| − | | style="background:#FFFFCC; color:black" align="center"| [[Image:VisualizationTutorial 3.6RC3.png| | + | | style="background:#FFFFCC; color:black" align="center" |[[Image:VisualizationTutorial 3.6RC3.png|250px]] |
|- | |- | ||
| − | | style="background:#FFFFCC; color:blue; font-size:110%" align="center"| | + | | id="ProgrammingInSlicer3Tutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Core''' |
| − | | style="background:#FFFFCC; color:black" valign="top"| ''' [[ | + | | style="background:#FFFFCC; color:black" valign="top" |''' [[Media:ProgrammingIntoSlicer3.6 SoniaPujol.pdf|Programming in Slicer3 Tutorial]]'''<br><br><br>The Programming in Slicer3 tutorial is an introduction to the integration of C++ stand-alone programs outside of the Slicer3 source tree.<br><br>'''Audience:''' Programmers and Engineers. |
| − | | style="background:#FFFFCC; color:black" valign="top" | [[Media: | + | | style="background:#FFFFCC; color:black" valign="top" |[[Media:HelloWorld Plugin.zip| '''HelloWorld Plugin''']]<br><br> The HelloWorld tutorial dataset contains an MR scan of the brain and pre-computed xml and C++ files for integrating the Hello Python plug-in to Slicer3. |
| − | | style="background:#FFFFCC; color:black" align="center"| [[Image:ProgrammingCourse_Logo.PNG| | + | | style="background:#FFFFCC; color:black" align="center" |[[Image:ProgrammingCourse_Logo.PNG|250px|Programming]] |
|- | |- | ||
| − | | style="background:#FFFFCC; color:blue; font-size:110%" align="center"| | + | | id="HelloPythonTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Core''' |
| − | | style="background:#FFFFCC; color:black" valign="top"|'''[[ | + | | style="background:#FFFFCC; color:black" valign="top" |''' [[Media:ProgrammingIntoSlicer3.6.1 HelloPython MICCAI2010 SoniaPujol.pdf|Hello Python Tutorial]]''' <br><br>The Hello Python tutorial is an introduction to the integration of Python stand-alone programs outside of the Slicer3 source tree.<br><br>'''Audience:''' Programmers and Engineers. |
| − | | style="background:#FFFFCC; color:black" valign="top" | | + | | style="background:#FFFFCC; color:black" valign="top" |[[Media:HelloPython.zip| '''HelloPython Plugin''']]<br><br> The HelloPython tutorial dataset contains an MR scan of the brain and pre-computed xml and Python code for integrating the Hello Python plug-in to Slicer3. |
| − | | style="background:#FFFFCC; color:black" align="center"| [[Image: | + | | style="background:#FFFFCC; color:black" align="center" |[[Image:HelloPythonTutorial.PNG|250px|Programming]] |
|- | |- | ||
| − | | style="background:#FFFFCC; color:blue; font-size:110%" align="center"| | + | | id="InteractiveEditor" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Core''' |
| − | | style="background:#FFFFCC; color:black" valign="top"|'''[[ | + | | style="background:#FFFFCC; color:black" valign="top" |'''[[Media:InteractiveEditorTutorial Slicer3.6-SoniaPujol.pdf|Interactive Editor]] '''<br><br><br>Shows how to use the interactive editing tools in Slicer. <br>'''Audience:''' All users and developers. |
| − | | style="background:#FFFFCC; color:black" valign="top" | '''[ | + | | style="background:#FFFFCC; color:black" valign="top" |'''[[Media:EditorTutorialDataset.zip|Editor Data]]'''<br><br>This dataset contains a MR dataset of the brain. |
| − | | style="background:#FFFFCC; color:black" align="center"| [[Image: | + | | style="background:#FFFFCC; color:black" align="center" |[[Image:InteractiveEditor.png|250px]] |
|- | |- | ||
| − | | style="background:#FFFFCC; color:blue; font-size:110%" align="center"| < | + | | id="ManualRegistration" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Core''' |
| − | | style="background:#FFFFCC; color:black" valign="top"| '''[[ | + | | style="background:#FFFFCC; color:black" valign="top" |'''[[Media:ManualRegistration Slicer3.6.pdf| Manual Registration]] ''' <br><br><br> Shows how to manually/interactively align two images in Slicer3.6 <br><br>'''Audience:''' First time & early users. |
| − | | style="background:#FFFFCC; color:black" valign="top" |''' [http://www.slicer.org/ | + | | style="background:#FFFFCC; color:black" valign="top" |'''[http://www.slicer.org/w/img_auth.php/8/88/Slicer3_Tutorial_ManualRegistration_ExampleDataset.zip Manual Registration Data]'''<br><br>This dataset contains two brain MRI of a single subject, obtained in different orientations. |
| − | | style="background:#FFFFCC; color:black" align="center"| [[Image:cc.PNG | | + | | style="background:#FFFFCC; color:black" align="center" |[[Image:Slicer3_ManualRegistrationTutorial.gif|250px]] |
| + | |- | ||
| + | | id="DiffusionMRITutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |'''[[Media:DiffusionMRITutorial UCSF2010 SoniaPujol.pdf| Diffusion MRI tutorial]]''' <br><br><br>This tutorial guides you through the process of loading diffusion MR data, estimating diffusion tensors, and performing tractography of white matter bundles. <br><br>'''Audience:''' All users and developers. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |'''[http://www.slicer.org/w/img_auth.php/c/cf/DiffusionDataset.zip Diffusion Data]''' | ||
| + | | style="background:#FFFFCC; color:black" align="center" |[[Image:cc.PNG |250px]] | ||
| + | |- | ||
| + | | id="ChangeTrackerTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |'''[[Media:Slicer3.6-ChangeTrackerTutorial.pdf| Change Tracker Tutorial]]''' <br><br><br>This tutorial describes the use of ChangeTracker module to detect changes in tumor volume from two MRI scans. <br><br>'''Audience:''' All users interested in longitudinal analysis of pathology. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[[Media:ChangeTracker.zip|Change Tracker Training Data]] Note: the dataset is also integrated with the ChangeTracker module (see Tutorial) | ||
| + | | style="background:#FFFFCC; color:black" align="center" |[[Image:Slicer3.4.1-ChangeTracker.jpg|250px]] | ||
| + | |- | ||
| + | | id="FreeSurferCourse" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |'''[[Media:FreeSurferCourse Slicer3.6 SoniaPujol.pdf| FreeSurfer Course]]''' The FreeSurfer dataset contains an MR scan of the brain and pre-computed FreeSurfer segmentation and cortical surface reconstructions. | ||
| + | <br><br>'''Audience:''' All users | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |'''[http://www.na-mic.org/Wiki/index.php/File:FreeSurferTutorialData.zip FreeSurfer tutorial data]''' | ||
| + | | style="background:#FFFFCC; color:black" align="center" |[[Image:FreeSurferTutorial.PNG|250px]] | ||
| + | |- | ||
| + | | id="NeurosurgicalPlanningTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[[Media:SlicerNeurosurgeryTutorial-3.6.1.pdf| '''Neurosurgical Planning Tutorial''']] <br><br> This tutorial takes the trainee through a complete workup for neurosurgical patient-specific mapping. Also see this tutorial for information on how to use Slicer's affine registration, simple region growing, model maker and tractography modules.<br> <br>'''Audience:''' All users interested in image-guided therapy. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[[Media:Slicer-tutorial-neurosurgery slicer3.6.1.zip| '''Neurosurgical Planning Data ''']] | ||
| + | | style="background:#FFFFCC; color:black" align="center" |[[Image:NeurosurgicalPlanningOverview.png|250px]] | ||
| + | |- | ||
| + | | id="PETCTSUVTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[http://www.na-mic.org/Wiki/index.php/File:PETCTTutorial.pptx '''PET/CT SUV Tutorial'''] <br><br> This tutorial takes the trainee through the computation of SUV body weight on a baseline and followup study. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[[Media:PETCTFusion-TutorialData.zip| PETCTFusion]] | ||
| + | | style="background:#FFFFCC; color:black" align="center" |[[Image:PETCTFusionBig.png|250px]] | ||
| + | |- | ||
| + | | id="EMSegmenterSimpleMode" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[[Media:EMSegmenterTutorialSimpleMode-Slicer-3.6.3-1.pdf| '''EMSegmenter Simple Mode ''']] <br><br> This tutorial takes the trainee through the segmentation of a MRI Human Brain without to adjust any parameters. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/c/cd/MRIHumanBrain_T1_aligned.nrrd '''Human Brain T1 Data'''] | ||
| + | | style="background:#FFFFCC; color:black" align="center" |[[Image:EMSegmenter-Simple-Mode.png|250px]] | ||
| + | |- | ||
| + | | id="EMSegmenterAdvancedMode" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[[Media:EMSegmenterTutorialAdvancedMode-Slicer-3.6.3-2.pdf| '''EMSegmenter Advanced Mode ''']] <br><br> This tutorial takes the trainee through the segmentation of a MRI Human Brain. The trainee will learn how to setup the EMSegmenter, this includes the creation of a task, the creation of an anatomical tree and adjusting the weights for the EM algorithm. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/c/cd/MRIHumanBrain_T1_aligned.nrrd '''Human Brain T1 Data'''] | ||
| + | | style="background:#FFFFCC; color:black" align="center" |[[Image:EMSegmenter-Advanced-Mode.png|250px]] | ||
| + | |- | ||
| + | | id="OpenIGTLink" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[[Media:OpenIGTLinkTutorial Slicer3.6.2 JunichiTokuda Dec2010.pdf| '''OpenIGTLink''']] <br><br> This tutorial shows to connect an IGT device using the OpenIGTLink. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[[Modules:OpenIGTLinkIF-3.6-Simulators'''OpenIGTLink|Data Simulator]] | ||
| + | | style="background:#FFFFCC; color:black" align="center" |[[Image:OpenIGT.PNG|250px]] | ||
| + | |- | ||
| + | | id="WhiteMatterExplorationForNeurosurgicalPlanning" style="background:#FFFFCC; color:blue; font-size:110%" align="center" |'''Workflow''' | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[[Media:WhiteMatterExplorationNeurosurgicalPlanning JHU2011 SoniaPujol.pdf| '''White Matter Exploration for Neurosurgical Planning''']] <br><br> This [[White_Matter_Exploration_Tutorial | tutorial]] walks the user through a workflow for exploring white matter fibers surrounding a tumor using Diffusion Tensor Imaging (DTI) Tractography. | ||
| + | | style="background:#FFFFCC; color:black" valign="top" |[[Media:WhiteMatterExplorationData.zip| White Matter Exploration dataset]] | ||
| + | | style="background:#FFFFCC; color:black" align="center" |[[Image:WhiteMatterExploration.PNG|250px]] | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | =Summer 2011 Tutorial Contest Entries (under construction)= | ||
| + | |||
| + | The following tutorials were part of the [http://wiki.na-mic.org/Wiki/index.php/Summer_2011_Tutorial_Contest Summer2011 Tutorial Contest]. | ||
| + | |||
| + | {| border="1" cellpadding="3" width="1000px" | ||
| + | |- style="background:#FFCC99; color:black; font-size:130%" align="center" | ||
| + | | style="width:100px" |'''Category''' | ||
| + | | style="width:350px" |'''Tutorial''' | ||
| + | | style="width:350px" |'''Sample Data''' | ||
| + | | style="width:250px" |'''Image''' | ||
| + | |- | ||
| + | | id="Automatic Segmentation of Traumatic Brain Injury MRI Volumes" style="background:#FFFFFF; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFFF; color:black" valign="top" |[[Media:TBISegmentation tutorial.pdf| '''Automatic Segmentation of Traumatic Brain Injury MRI Volumes''']] <br><br> This [http://www.na-mic.org/Wiki/index.php/TBISegmentation_TutorialContestSummer2011 tutorial] guides the user through the automatic segmentation of TBI volumes using Atlas Based classification. | ||
| + | | style="background:#FFFFFF; color:black" valign="top" |[[Media:TBISegmentation data.zip| TBI segmentation dataset]] | ||
| + | | style="background:#FFFFF; color:black" align="center" |[[Image:TBI_image.png|250px]] | ||
| + | |- | ||
| + | | id="Centerline Extraction of Coronary Arteries" style="background:#FFFFFF; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFFF; color:black" valign="top" |[http://wiki.na-mic.org/Wiki/images/a/aa/VMTK_Slicer_3.6.3_Updated_OCT_2011.pdf '''Centerline Extraction of Coronary Arteries using VMTK in 3D Slicer'''] <br><br> This [http://wiki.na-mic.org/Wiki/images/a/aa/VMTK_Slicer_3.6.3_Updated_OCT_2011.pdf tutorial] guides the user step by step through the process of centerline extraction of Coronary Arteries in a cardiac blood-pool MRI using VMTK based Tools. | ||
| + | | style="background:#FFFFFF; color:black" valign="top" |[http://www.na-mic.org/Wiki/index.php/File:TutorialVMTKCoronariesCenterlinesMRI_Data_Winter2010AHM.zip Cardiac blood-pool MRI dataset] | ||
| + | | style="background:#FFFFF; color:black" align="center" |[[Image:Vmtkcloseupvoronoicenterlinewithreference.png|250px]] | ||
|- | |- | ||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
| − | =Summer 2010 Tutorial Contest Entries | + | =Summer 2010 Tutorial Contest Entries= |
| − | + | ||
| + | The following tutorials were part of the [http://wiki.na-mic.org/Wiki/index.php/Summer_2010_Tutorial_Contest#List_of_submitted_tutorials Summer 2010 Slicer Tutorial Contest]. | ||
| − | {| border="1" cellpadding=" | + | {| border="1" cellpadding="3" width="1000px" |
|- style="background:#FFCC99; color:black; font-size:130%" align="center" | |- style="background:#FFCC99; color:black; font-size:130%" align="center" | ||
| − | |'''Category''' | + | | style="width:100px" |'''Category''' |
| − | |'''Tutorial''' | + | | style="width:350px" |'''Tutorial''' |
| − | |'''Sample Data''' | + | | style="width:350px" |'''Sample Data''' |
| − | |'''Image''' | + | | style="width:250px" |'''Image''' |
| + | |- | ||
| + | | id="IAFEMesh" style="background:#FFFFFF ; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFFF color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/1/13/IAFEMesh-TutorialContestSummer2010.pdf Meshing Workflow] <br><br> '''Audience:''' All users and developers. (Note: Mac and Linux only.) | ||
| + | | style="background:#FFFFFF ; color:black" valign="top" |[[File:IAFEMeshData-TutorialContestSummer2010.zip|Meshing Workflow Data]]<br><br> | ||
| + | | style="background:#FFFFFF ; color:black" align="center" |[[File:FEMesh.png |250px]] | ||
| + | |- | ||
| + | | id="Fiducials" style="background:#FFFFFF ; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFFF ; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/6/66/Fiducials_TutorialContestSummer2010.pdf Fiducials] <br><br>'''Audience:''' All users and developers. | ||
| + | | style="background:#FFFFFF ; color:black" valign="top" |[http://wiki.na-mic.org/Wiki/index.php/File:Fiducials_TutorialContestSummer2010.zip Fiducials Data]<br><br> | ||
| + | | style="background:#FFFFFF ; color:black" align="center" |[[File:Fiducials.png |250px]] | ||
| + | |- | ||
| + | | id="RobustStatisticSegmenter" style="background:#FFFFFF ; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFFF ; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/3/3c/RSS_TutorialContestSummer2010.pdf Robust Statistic Segmenter] <br><br>'''Audience:''' All users and developers. | ||
| + | | style="background:#FFFFFF ; color:black" valign="top" |[http://wiki.na-mic.org/Wiki/index.php/File:RSSData_TutorialContestSummer2010.zip Robust Statistic Segmenter Data]<br><br> | ||
| + | | style="background:#FFFFFF ; color:black" align="center" |[[File:RSS.png |250px]] | ||
| + | |- | ||
| + | | id="LongitudinalLesionComparison" style="background:#FFFFFF ; color:blue; font-size:110%" align="center" |'''Specialized''' | ||
| + | | style="background:#FFFFFF ; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/8/8c/Longitudinal_Lesion_Comparison_TutorialContest_2010.pdf Longitudinal Lesion Comparison] <br><br>'''Audience:''' All users and developers. (Note: Mac and Linux only.) | ||
| + | | style="background:#FFFFFF ; color:black" valign="top" |[http://wiki.na-mic.org/Wiki/index.php/File:LongitudinalLesionComparison2_TutorialContestSummer2010.zip Longitudinal Lesion Comparison Data] | ||
| + | | style="background:#FFFFFF ; color:black" align="center" |[[File:Longitudinal_Lesion.png |250px]] | ||
|- | |- | ||
| − | | style="background:#FFFFFF ; color:blue; font-size:110%" align="center"| | + | | id="ProstateNav" style="background:#FFFFFF ; color:blue; font-size:110%" align="center" |'''Specialized''' |
| − | | style="background:#FFFFFF | + | | style="background:#FFFFFF ; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/0/06/ProstateNav_TutorialContestSummer2010.pdf Robot-Assisted MRI-Guided Prostate Biopsy] <br><br>'''Audience:''' All users and developers. |
| − | | style="background:#FFFFFF ; color:black" valign="top" | [http://www.slicer.org/ | + | | style="background:#FFFFFF ; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/3/3a/ProstateNavData_TutorialContestSummer2010.zip Robot-Assisted MRI-Guided Prostate Biopsy] |
| − | | style="background:#FFFFFF ; color:black" align="center"| | + | | style="background:#FFFFFF ; color:black" align="center" |[[File:ProstateNav.png |250px]] |
|- | |- | ||
| − | | style="background:#FFFFFF ; color:blue; font-size:110%" align="center"| | + | | id="LabelFusion" style="background:#FFFFFF ; color:blue; font-size:110%" align="center" |'''Specialized''' |
| − | | style="background:#FFFFFF ; color:black" valign="top"| [http:// | + | | style="background:#FFFFFF ; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/0/0a/LabelFusion_Tutorial.pdf Atlas Label Fusion & Surface Registration] <br><br>'''Audience:''' All users and developers. |
| − | | style="background:#FFFFFF ; color:black" valign="top" | [http:// | + | | style="background:#FFFFFF ; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/2/27/AtlasLabelFusion_TutorialContestSummer2010.zip Atlas Label Fusion & Surface Registration] |
| − | | style="background:#FFFFFF ; color:black" align="center"| | + | | style="background:#FFFFFF ; color:black" align="center" |[[File:AtlasLabelFusion.png |250px]] |
|- | |- | ||
| − | | style="background:#FFFFFF ; color:blue; font-size:110%" align="center"| | + | | id="Stochastic_Tractography" style="background:#FFFFFF ; color:blue; font-size:110%" align="center" |'''Specialized''' |
| − | | style="background:#FFFFFF ; color:black" valign="top"| [http:// | + | | style="background:#FFFFFF ; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/9/90/Stochastic_Tractography_TutorialContestSummer2010.pdf Stochastic Tractography] <br><br>'''Audience:''' All users and developers. (Note: Mac and Linux only.) |
| − | | style="background:#FFFFFF ; color:black" valign="top" | [http:// | + | | style="background:#FFFFFF ; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/7/70/Stochastic_tutorial_data_TutorialContestSummer2010.zip Stochastic Tractography] |
| − | | style="background:#FFFFFF ; color:black" align="center"| | + | | style="background:#FFFFFF ; color:black" align="center" |[[File:Stochastic_tutorial_uncinate.JPG |250px]] |
|- | |- | ||
| − | | style="background:#FFFFFF ; color:blue; font-size:110%" align="center"| | + | | id="PerkStationModule" style="background:#FFFFFF ; color:blue; font-size:110%" align="center" |'''Specialized''' |
| − | | style="background:#FFFFFF ; color:black" valign="top"|[ | + | | style="background:#FFFFFF ; color:black" valign="top" |[[File:PerkStationModule_TutorialContestSummer2010.pdf|PERK Station Module]] <br><br>'''Audience:''' All users and developers. (Note: Windows only.) |
| − | | style="background:#FFFFFF ; color:black" valign="top" | [http:// | + | | style="background:#FFFFFF ; color:black" valign="top" |[http://www.slicer.org/w/img_auth.php/3/36/PerkStationData_TutorialContestSummer2010.zip PERK Station Module] |
| − | | style="background:#FFFFFF ; color:black" align="center"| | + | | style="background:#FFFFFF ; color:black" align="center" |[[File:PERKStation.png |250px]] |
|- | |- | ||
|} | |} | ||
=Software Installation= | =Software Installation= | ||
| + | |||
*The [http://www.slicer.org/pages/Special:SlicerDownloads '''Slicer download page'''] contains information on how to obtain a compiled version of Slicer for a variety of platforms and where to find the source code for Slicer 3. | *The [http://www.slicer.org/pages/Special:SlicerDownloads '''Slicer download page'''] contains information on how to obtain a compiled version of Slicer for a variety of platforms and where to find the source code for Slicer 3. | ||
=Software Documentation= | =Software Documentation= | ||
| + | |||
*For the Slicer 3.6 manual pages please click [[Documentation-3.6|here]]. These pages are the reference manual for Slicer 3.6 and briefly explain the functionality found in panels and modules. | *For the Slicer 3.6 manual pages please click [[Documentation-3.6|here]]. These pages are the reference manual for Slicer 3.6 and briefly explain the functionality found in panels and modules. | ||
| + | |||
| + | =External Resources= | ||
| + | |||
| + | This set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field: | ||
| + | |||
| + | *[http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial.html Open Source Paleontologist: 3D Slicer: The Tutorial] | ||
| + | *[http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial-part-ii.html Open Source Paleontologist: 3D Slicer: The Tutorial Part II] | ||
| + | *[http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial-part-iii.html Open Source Paleontologist: 3D Slicer: The Tutorial Part III] | ||
| + | *[http://openpaleo.blogspot.com/2008/12/3d-slicer-tutorial-part-iv.html Open Source Paleontologist: 3D Slicer: The Tutorial Part IV] | ||
| + | *[http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-v.html Open Source Paleontologist: 3D Slicer: The Tutorial Part V] | ||
| + | *[http://openpaleo.blogspot.com/2009/03/3d-slicer-tutorial-part-vi.html Open Source Paleontologist: 3D Slicer: The Tutorial Part VI] | ||
| + | |||
| + | Narrated screencast video showing how to load data and make models: http://www.youtube.com/watch?v=GNsWRnm7gQw&context=C42d9859ADvjVQa1PpcFPhXz87xj_xyVq19zSUexfAbEm8Iz3DYEk= | ||
| + | |||
| + | |||
| + | *[http://www.imm.dtu.dk/projects/BRATS2012/Jakab_TumorSegmentation_Manual.pdf Brain Tumor Segmentation Manual (Slicer 3.6)] | ||
Latest revision as of 14:24, 28 January 2020
Home < Slicer 3.6:TrainingContents
Slicer 3.6 Tutorials
- The following table contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version 3.6 release) to accomplish certain tasks.
- For tutorials for other versions of Slicer, please visit the Slicer training portal.
- For questions related to the Slicer3 Compendium, please send an e-mail to Sonia Pujol, Ph.D
| Category | Tutorial | Sample Data | Image |
| Core | Slicer3Minute Tutorial The Slicer3Minute tutorial is an introduction to the advanced 3D visualization capabilities of Slicer3.6. Audience: First time users. |
Slicer3Minute Data The Slicer3Minute dataset contains an MR scan of the brain and 3D reconstructions of the anatomy |
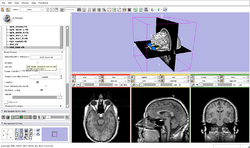
|
| Core | Slicer3Visualization Tutorial The Slicer3Visualization tutorial guides through 3D data loading and visualization in Slicer3.6. Audience: All beginners including clinicians, scientists, engineers and programmers. |
Slicer3Visualization Data The Slicer3VisualizationDataset contains two MR scans of the brain, a pre-computed labelmap and 3D reconstructions of the anatomy. |
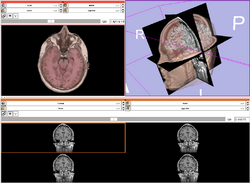
|
| Core | Programming in Slicer3 Tutorial The Programming in Slicer3 tutorial is an introduction to the integration of C++ stand-alone programs outside of the Slicer3 source tree. Audience: Programmers and Engineers. |
HelloWorld Plugin The HelloWorld tutorial dataset contains an MR scan of the brain and pre-computed xml and C++ files for integrating the Hello Python plug-in to Slicer3. |
|
| Core | Hello Python Tutorial The Hello Python tutorial is an introduction to the integration of Python stand-alone programs outside of the Slicer3 source tree. Audience: Programmers and Engineers. |
HelloPython Plugin The HelloPython tutorial dataset contains an MR scan of the brain and pre-computed xml and Python code for integrating the Hello Python plug-in to Slicer3. |
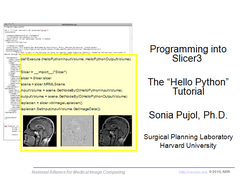
|
| Core | Interactive Editor Shows how to use the interactive editing tools in Slicer. Audience: All users and developers. |
Editor Data This dataset contains a MR dataset of the brain. |
|
| Core | Manual Registration Shows how to manually/interactively align two images in Slicer3.6 Audience: First time & early users. |
Manual Registration Data This dataset contains two brain MRI of a single subject, obtained in different orientations. |
|
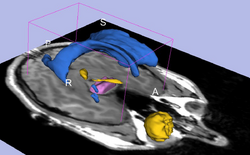
| Specialized | Diffusion MRI tutorial This tutorial guides you through the process of loading diffusion MR data, estimating diffusion tensors, and performing tractography of white matter bundles. Audience: All users and developers. |
Diffusion Data | 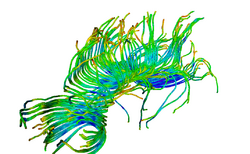
|
| Specialized | Change Tracker Tutorial This tutorial describes the use of ChangeTracker module to detect changes in tumor volume from two MRI scans. Audience: All users interested in longitudinal analysis of pathology. |
Change Tracker Training Data Note: the dataset is also integrated with the ChangeTracker module (see Tutorial) | |
| Specialized | FreeSurfer Course The FreeSurfer dataset contains an MR scan of the brain and pre-computed FreeSurfer segmentation and cortical surface reconstructions.
|
FreeSurfer tutorial data | 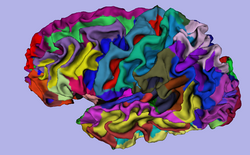
|
| Specialized | Neurosurgical Planning Tutorial This tutorial takes the trainee through a complete workup for neurosurgical patient-specific mapping. Also see this tutorial for information on how to use Slicer's affine registration, simple region growing, model maker and tractography modules. Audience: All users interested in image-guided therapy. |
Neurosurgical Planning Data | 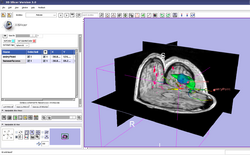
|
| Specialized | PET/CT SUV Tutorial This tutorial takes the trainee through the computation of SUV body weight on a baseline and followup study. |
PETCTFusion | 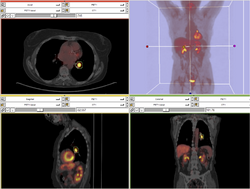
|
| Specialized | EMSegmenter Simple Mode This tutorial takes the trainee through the segmentation of a MRI Human Brain without to adjust any parameters. |
Human Brain T1 Data | 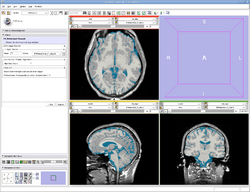
|
| Specialized | EMSegmenter Advanced Mode This tutorial takes the trainee through the segmentation of a MRI Human Brain. The trainee will learn how to setup the EMSegmenter, this includes the creation of a task, the creation of an anatomical tree and adjusting the weights for the EM algorithm. |
Human Brain T1 Data | 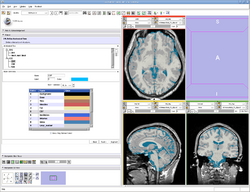
|
| Specialized | OpenIGTLink This tutorial shows to connect an IGT device using the OpenIGTLink. |
Data Simulator | 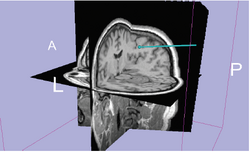
|
| Workflow | White Matter Exploration for Neurosurgical Planning This tutorial walks the user through a workflow for exploring white matter fibers surrounding a tumor using Diffusion Tensor Imaging (DTI) Tractography. |
White Matter Exploration dataset | 
|
Summer 2011 Tutorial Contest Entries (under construction)
The following tutorials were part of the Summer2011 Tutorial Contest.
| Category | Tutorial | Sample Data | Image |
| Specialized | Automatic Segmentation of Traumatic Brain Injury MRI Volumes This tutorial guides the user through the automatic segmentation of TBI volumes using Atlas Based classification. |
TBI segmentation dataset | 
|
| Specialized | Centerline Extraction of Coronary Arteries using VMTK in 3D Slicer This tutorial guides the user step by step through the process of centerline extraction of Coronary Arteries in a cardiac blood-pool MRI using VMTK based Tools. |
Cardiac blood-pool MRI dataset | 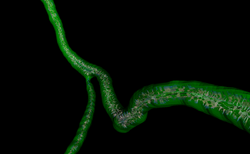
|
Summer 2010 Tutorial Contest Entries
The following tutorials were part of the Summer 2010 Slicer Tutorial Contest.
| Category | Tutorial | Sample Data | Image |
| Specialized | Meshing Workflow Audience: All users and developers. (Note: Mac and Linux only.) |
File:IAFEMeshData-TutorialContestSummer2010.zip |
|
| Specialized | Fiducials Audience: All users and developers. |
Fiducials Data |
|
| Specialized | Robust Statistic Segmenter Audience: All users and developers. |
Robust Statistic Segmenter Data |
|
| Specialized | Longitudinal Lesion Comparison Audience: All users and developers. (Note: Mac and Linux only.) |
Longitudinal Lesion Comparison Data | 
|
| Specialized | Robot-Assisted MRI-Guided Prostate Biopsy Audience: All users and developers. |
Robot-Assisted MRI-Guided Prostate Biopsy | 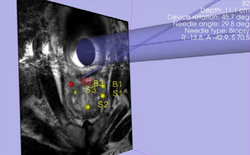
|
| Specialized | Atlas Label Fusion & Surface Registration Audience: All users and developers. |
Atlas Label Fusion & Surface Registration | 
|
| Specialized | Stochastic Tractography Audience: All users and developers. (Note: Mac and Linux only.) |
Stochastic Tractography | 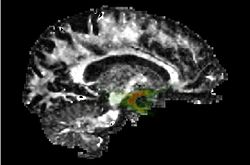
|
| Specialized |  Audience: All users and developers. (Note: Windows only.) |
PERK Station Module | 
|
Software Installation
- The Slicer download page contains information on how to obtain a compiled version of Slicer for a variety of platforms and where to find the source code for Slicer 3.
Software Documentation
- For the Slicer 3.6 manual pages please click here. These pages are the reference manual for Slicer 3.6 and briefly explain the functionality found in panels and modules.
External Resources
This set of tutorials about the use of slicer in paleontology is very well written and provides step-by-step instructions. Even though it covers slicer version 3.4, many of the concepts and techniques have applicability to the new version and to any 3D imaging field:
- Open Source Paleontologist: 3D Slicer: The Tutorial
- Open Source Paleontologist: 3D Slicer: The Tutorial Part II
- Open Source Paleontologist: 3D Slicer: The Tutorial Part III
- Open Source Paleontologist: 3D Slicer: The Tutorial Part IV
- Open Source Paleontologist: 3D Slicer: The Tutorial Part V
- Open Source Paleontologist: 3D Slicer: The Tutorial Part VI
Narrated screencast video showing how to load data and make models: http://www.youtube.com/watch?v=GNsWRnm7gQw&context=C42d9859ADvjVQa1PpcFPhXz87xj_xyVq19zSUexfAbEm8Iz3DYEk=