Difference between revisions of "Modules:LesionSegmentationApplications-Documentation-3.6"
(Intital page) |
(→Authors, Collaborators & Contact: Updated email addr.) |
||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 9: | Line 9: | ||
{| | {| | ||
|[[File:LesionFlowChart.jpg|thumb|200px|Lesion Segmentation Method Overview]] | |[[File:LesionFlowChart.jpg|thumb|200px|Lesion Segmentation Method Overview]] | ||
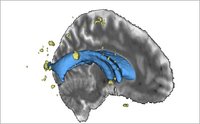
| + | |[[File:LesionsWithVentricles.png|thumb|200px|Segmented Lesion Volume in Brain]] | ||
|} | |} | ||
| Line 19: | Line 20: | ||
===Authors, Collaborators & Contact=== | ===Authors, Collaborators & Contact=== | ||
| − | * Author1: Mark Scully, The | + | * Author1: Mark Scully, The University of Iowa |
* Author2: H. J. Bockholt, The Mind Research Network | * Author2: H. J. Bockholt, The Mind Research Network | ||
* Contributor1: | * Contributor1: | ||
| − | * Contact: Mark Scully, | + | * Contact: Mark Scully, mark-scully[at]uiowa[dot]edu |
===Module Description=== | ===Module Description=== | ||
The lupus lesion segmentation applications are a group of tools for segmenting lesions in lupus patients using the T1-weighted, T2-weighted, and FLAIR images. This cross-platform tool can be run within Slicer3 as an external module, or directly as a command line. | The lupus lesion segmentation applications are a group of tools for segmenting lesions in lupus patients using the T1-weighted, T2-weighted, and FLAIR images. This cross-platform tool can be run within Slicer3 as an external module, or directly as a command line. | ||
| + | |||
| + | Please see the [http://wiki.na-mic.org/Wiki/index.php/DBP2:MIND:Roadmap DBP description] for more information. | ||
== Usage == | == Usage == | ||
| Line 49: | Line 52: | ||
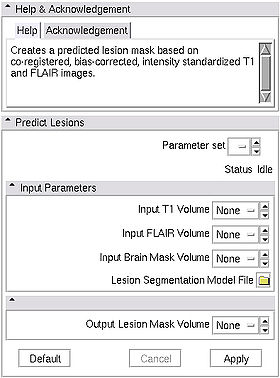
** Input Brain Mask Volume: set input Brain Mask image, if available | ** Input Brain Mask Volume: set input Brain Mask image, if available | ||
** Lesion segmentation Model File: set to the model file available with the tutorial | ** Lesion segmentation Model File: set to the model file available with the tutorial | ||
| − | |||
* '''Output:''' set output image | * '''Output:''' set output image | ||
| Line 96: | Line 98: | ||
===References=== | ===References=== | ||
| − | + | Scully M, Anderson B, Lane T, Gasparovic C, Magnotta V, Sibbitt W, Roldan C, Kikinis R and Bockholt HJ (2010) An automated method for segmenting white matter lesions through multi-level morphometric feature classification with application to lupus. Front. Hum. Neurosci. doi:10.3389/fnhum.2010.00027 | |
| + | http://frontiersin.org/neuroscience/humanneuroscience/paper/10.3389/fnhum.2010.00027/ | ||
Latest revision as of 19:24, 10 November 2011
Home < Modules:LesionSegmentationApplications-Documentation-3.6Return to Slicer 3.6 Documentation
Lupus Lesion Segmentation Applications
General Information
Module Type & Category
Type: CLI
Category: Segmentation
Authors, Collaborators & Contact
- Author1: Mark Scully, The University of Iowa
- Author2: H. J. Bockholt, The Mind Research Network
- Contributor1:
- Contact: Mark Scully, mark-scully[at]uiowa[dot]edu
Module Description
The lupus lesion segmentation applications are a group of tools for segmenting lesions in lupus patients using the T1-weighted, T2-weighted, and FLAIR images. This cross-platform tool can be run within Slicer3 as an external module, or directly as a command line.
Please see the DBP description for more information.
Usage
Use Cases, Examples
This module is specifically for segmenting lesions in lupus patients.
Tutorials
- Tutorials:
- Tutorial Dataset
Quick Tour of Features and Use
|
Development
Notes from the Developer(s)
Algorithms used, library classes depended upon, use cases, etc.
Dependencies
- Slicer3 modules:
Tests
On the Dashboard, these tests verify that the module is working on various platforms:
- MyModuleTest1 MyModuleTest1.cxx
- MyModuleTest2 MyModuleTest2.cxx
Known bugs
Links to known bugs in the Slicer3 bug tracker
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
- Source code
- Download release source code via svn:
- cvs -d :pserver:anonymous@www.nitrc.org:/cvsroot/lupuslesion login
- cvs -d :pserver:anonymous@www.nitrc.org:/cvsroot/lupuslesion checkout lupuslesion
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from http://nihroadmap.nih.gov/bioinformatics.
References
Scully M, Anderson B, Lane T, Gasparovic C, Magnotta V, Sibbitt W, Roldan C, Kikinis R and Bockholt HJ (2010) An automated method for segmenting white matter lesions through multi-level morphometric feature classification with application to lupus. Front. Hum. Neurosci. doi:10.3389/fnhum.2010.00027 http://frontiersin.org/neuroscience/humanneuroscience/paper/10.3389/fnhum.2010.00027/