Difference between revisions of "Modules:ChangeTracker-Documentation-3.4"
| Line 115: | Line 115: | ||
===Known bugs=== | ===Known bugs=== | ||
| + | |||
| + | This module isn't fully integrated with MRML yet. MRML scene snapshots and restores are not yet supported. Please save your work using the file save dialogs in the user interface. | ||
Follow this [http://na-mic.org/Mantis/main_page.php link] to the Slicer3 bug tracker. | Follow this [http://na-mic.org/Mantis/main_page.php link] to the Slicer3 bug tracker. | ||
Revision as of 13:55, 5 March 2009
Home < Modules:ChangeTracker-Documentation-3.4Return to Slicer 3.4 Documentation
ChangeTracker
General Information
Module Type & Category
Type: Interactive
Category: Wizards
Authors, Collaborators & Contact
- Kilian Pohl, IBM Research/SPL
- Ender Konukoglu, INRIA
- Andriy Fedorov, SPL
- Ron Kikinis, SPL
- Contact: Andriy Fedorov, fedorov at spl dot bwh dot edu
Module Description
ChangeTracker is a software tool for quantification of the subtle changes in pathology. The module provides a workflow pipeline that combines user input with the medical data. As a result we provide quantitative volumetric measurements of growth/shrinkage together with the volume rendering of the tumor and color-coded visualization of the tumor growth/shrinkage.
Usage
Examples, Use Cases & Tutorials
- The module has been designed and tested for measuring meningioma development.
- Detecting subtle change in pathology tutorial: slides, data
Quick Tour of Features and Use
ChangeTracker uses workflow processing of the input data guided by minimal user interaction. There are 4 processing steps.
- Step 1: Define input scans
| Use drop-down controls to choose the two scans where you would like to measure pathology development. Currently, we support analysis of the images that correspond to two time points. Extending of the module to accept more than two time points is the feature under development.
Note, that ChangeTracker does not "see" transforms located above volume nodes in the MRML hierarchy. If you want any transform in the MRML tree to be used as the initial pose of the image, you will need to create a new volume by resampling the original volume. Slicer3 module ResampleVolume2 can be used for this purpose. |
- Step 2: Define volume of interest
This step of wizard includes the following user cotrols to facilitate Volume of Interest (VOI) selection:
- "Hide/show render" button: used to control visibility of volume rendering for the selected region
- "Reset" button: reset the selected region to start over
- "ROI Widget Controls: RAS Space" frame: contains sliders to initialize VOI in RAS (physical) space
- "ROI Widget Controls: IJK Space" frame: contains sliders to initialize VOI in IJK (voxel) space
"ROI Widget" refers to the three-dimensional selection box that appears in the 3d slice view once you begin to select VOI.
You can define VOI using either one or combination of the following methods:
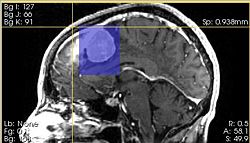
- Red/Yellow/Green Slice view bounding box: click left mouse button to define the coordinates of the bounding box that encloses analyzed target in the image. Volume of interest is highlighted in the slice views by semi-transparent blue overlay.
- 3d Slice view: use the ROI Widget controls to resize and/or drag the VOI
- Slider controls: use the slider controls in the wizard frame to resize or move VOI
Note, that the last method is recommended only if you want to enter the VOI parameters you already know. This method is not practical to identify VOI in a new image.
- Step 3: Select tumor segmentation threshold
| Use threshold control slider to find the intensity that most closely approximates tumor volume. Thresholded volume is rendered interactively in the 3D viewer as you are adjusting the threshold value, and is also visualized as semi-transparent label in the image slice viewers.
Note, that currently ChangeTracker expects that the tissue you monitor corresponds to the high part of the image intensity distribution. |
- Step 4: Define metric
| We provide two metrics to quantify tumor progression: intensity-based and deformation-based (see details in this paper). Choose the metric(s) you would like to use. |
- Analysis/Results
| Results are reported as the change in tumor volume, separately for growth and shrinkage component. The quantitative results are reported in voxels and mm^3. Sensitivity of the analysis can be controlled by choosing Sensitive, Moderate, or Robust analysis approach (Robust being the most conservative). As you change the sensitivity, visualization and quantitative data are updated accordingly. Growth is shown in purple, and shrinkage -- in green color.
The visualization of the analysis results includes the following components upon the completion of analysis:
You can choose to display the original sampling grid, and view the coronal slice of the subject using the wizard controls. |
Development
Dependencies
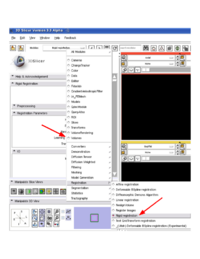
ChangeTracker depends on the following modules: Volumes, Rigid Registration, DiffeomorphicDemons
|
Volumes module is required for the general tasks of new volume creation etc. This module should always be available in your Slicer, but can be turned off in the Slicer Application Settings. Make sure it is available. ChangeTracker is using Rigid Registration module through the CommandLineModule shared object invocation. Note, that there are two distinct rigid registration modules in Slicer, which have identical functionality: "Linear registration" and "Rigid registration". Both of these modules should be available in your build, but "Rigid registration" *must* be available for ChangeTracker to function properly. You can verify the availability of the module manually by looking at the list of modules in the registration category. |
Known bugs
This module isn't fully integrated with MRML yet. MRML scene snapshots and restores are not yet supported. Please save your work using the file save dialogs in the user interface.
Follow this link to the Slicer3 bug tracker.
Currently unresolved bugs:
Usability issues
| ChangeTracker is using rigid registration module to automatically align the input images and selected volumes of interest. It is possible, that rigid registration can fail to align your data. If this happens, you will see error message in the pop-up box, as shown to the right.
You can try to solve this problem by manually registering your data prior to using it with ChangeTracker. You can try to register your data by using the Rigid registration module and manually adjusting the registration parameters to have satisfactory result of registration. Note, that you will need to select "Create new volume" while running registration, and use that volume together with the fixed image as inputs to ChangeTracker. In the case if Rigid registration module fails to align your data, you can use Transforms module to perform manual registration of your data. Follow this link to the Slicer3 bug tracker to report any usability issues. Please select the usability issue category when browsing or contributing. |
Source code & documentation
Follow this link to the ChangeTracker source code in ViewVC.
Documentation generated by doxygen.
More Information
Acknowledgment
ChangeTracker development has been funded by Brain Science Foundation
References
- E.Konukoglu, W.M.Wells, S.Novellas, N.Ayache, R.Kikinis, P.M.Black, K.M.Pohl. Monitoring Slowly Evolving Tumors. Proc. of 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro 2008, pp.812-815 link