Difference between revisions of "Modules:ChangeTracker-Documentation"
| (7 intermediate revisions by the same user not shown) | |||
| Line 39: | Line 39: | ||
* '''Step 1: Define input scans''' | * '''Step 1: Define input scans''' | ||
| − | + | {| | |
| − | + | |Use drop-down controls to choose the two scans where you would like to measure pathology development. Currently, we support analysis of the images that correspond to two time points. Extending of the module to accept more than two time points is the feature under development. | |
| − | Image: | + | |[[Image:ChangeTracker_Step1.png|thumb|250px|Step 1: Scan selection]] |
| − | + | |} | |
| + | |||
| + | * '''Step 2: Define volume of interest''' | ||
{| | {| | ||
| + | |Use the mouse pointer in combination with left button clicks to identify the bounding box enclosing pathology in three projections of the viewer. You can also use the mouse to adjust sliders defining the bounding box more precisely. If you made a mistake, click "Reset" button to start over. "Show VOI" button can be used to show/hide blue overlay of the region of interest of the the image slice views. | ||
| + | |{| | ||
|[[Image:ChangeTracker_Step2_1.png|thumb|200px|Step 2: ROI selection]] | |[[Image:ChangeTracker_Step2_1.png|thumb|200px|Step 2: ROI selection]] | ||
| − | |[[Image:ChangeTracker_Step2_2.png|thumb| | + | |[[Image:ChangeTracker_Step2_2.png|thumb|250px|Step 2: ROI visualization]] |
|} | |} | ||
| + | |||
* '''Step 3: Select tumor segmentation threshold''' | * '''Step 3: Select tumor segmentation threshold''' | ||
| − | Use threshold control slider to find the intensity that most closely approximates tumor volume. Tumor volume is rendered in the 3D viewer as you are choosing the appropriate threshold. | + | {| |
| − | + | |Use threshold control slider to find the intensity that most closely approximates tumor volume. Tumor volume is rendered in the 3D viewer as you are choosing the appropriate threshold, and is also visualized as semi-transparent label in the image slice viewers. | |
| − | [[Image:ChangeTracker_Step3.png|thumb|450px|Step 3: Threshold selection]] | + | |[[Image:ChangeTracker_Step3.png|thumb|450px|Step 3: Threshold selection]] |
| + | |} | ||
* '''Step 4: Define metric''' | * '''Step 4: Define metric''' | ||
| − | We provide two metrics -- intensity-based and deformation-based -- to quantify tumor progression (see details in this [http://www.spl.harvard.edu/pages/Special:PubDB_View?dspaceid=1430 paper]). Choose the metric you would like to use. | + | {| |
| − | + | |We provide two metrics -- intensity-based and deformation-based -- to quantify tumor progression (see details in this [http://www.spl.harvard.edu/pages/Special:PubDB_View?dspaceid=1430 paper]). Choose the metric you would like to use. | |
| − | [[Image:ChangeTracker_Step4.png|thumb| | + | |[[Image:ChangeTracker_Step4.png|thumb|250px|Step 4: Metric selection]] |
| + | |} | ||
* '''Analysis/Results''' | * '''Analysis/Results''' | ||
| − | Results are reported as the change in tumor volume, separately for growth and shrinkage component. The quantitative results are reported in voxels and mm^3. | + | {| |
| + | |Results are reported as the change in tumor volume, separately for growth and shrinkage component. The quantitative results are reported in voxels and mm^3. | ||
Sensitivity of the analysis can be controlled by choosing Sensitive, Moderate, or Robust analysis approach (Robust being the most conservative). As you change the sensitivity, visualization and quantitative data are updated accordingly. Growth is shown in purple, and shrinkage -- in green color. | Sensitivity of the analysis can be controlled by choosing Sensitive, Moderate, or Robust analysis approach (Robust being the most conservative). As you change the sensitivity, visualization and quantitative data are updated accordingly. Growth is shown in purple, and shrinkage -- in green color. | ||
| Line 69: | Line 77: | ||
You can choose to display the original sampling grid, and view the coronal slice of the subject using the module controls. | You can choose to display the original sampling grid, and view the coronal slice of the subject using the module controls. | ||
| − | [[Image:ChangeTracker_Analysis.png|thumb|450px|Visualization of the analysis results]] | + | |[[Image:ChangeTracker_Analysis.png|thumb|450px|Visualization of the analysis results]] |
| + | |} | ||
== Development == | == Development == | ||
| Line 75: | Line 84: | ||
===Dependencies=== | ===Dependencies=== | ||
| − | + | {| | |
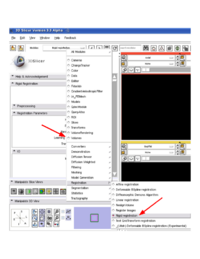
| + | |ChangeTracker is using [http://slicer.spl.harvard.edu/slicerWiki/index.php/Modules:LinearRegistration-Documentation Rigid Registration module] through the CommandLineModule shared object invocation. Note, that there are two distinct rigid registration modules in Slicer, which have identical functionality: "Linear registration" and "Rigid registration". Both of these modules should be available in your build, but "Rigid registration" *must* be available for ChangeTracker to function properly. You can verify the availability of the module manually by looking at the list of modules in the registration category. | ||
| + | |[[Image:rigid_registration_module.png|thumb|200px|Checking availability of Rigid registration module (click to zoom in)]] | ||
| + | |} | ||
===Known bugs=== | ===Known bugs=== | ||
Latest revision as of 23:51, 22 December 2008
Home < Modules:ChangeTracker-DocumentationReturn to Slicer Documentation
ChangeTracker
General Information
Module Type & Category
Type: Interactive
Category: Base/Segmentation
Authors, Collaborators & Contact
- Kilian Pohl, IBM Research/SPL
- Ender Konukoglu, INRIA
- Andriy Fedorov, SPL
- Ron Kikinis, SPL
- Contact: Andriy Fedorov, fedorov at spl dot bwh dot edu
Module Description
ChangeTracker is a software tool for quantification of the subtle changes in pathology. The module provides a workflow pipeline that combines user input with the medical data. As a result we provide quantitative volumetric measurements of growth/shrinkage together with the volume rendering of the tumor and color-coded visualization of the tumor growth/shrinkage.
Usage
Examples, Use Cases & Tutorials
- The module has been designed and tested for measuring meningioma development.
- Detecting subtle change in pathology tutorial: slides, data
Quick Tour of Features and Use
ChangeTracker uses workflow processing of the input data guided by minimal user interaction. There are 4 processing steps.
- Step 1: Define input scans
| Use drop-down controls to choose the two scans where you would like to measure pathology development. Currently, we support analysis of the images that correspond to two time points. Extending of the module to accept more than two time points is the feature under development. |
- Step 2: Define volume of interest
| Use the mouse pointer in combination with left button clicks to identify the bounding box enclosing pathology in three projections of the viewer. You can also use the mouse to adjust sliders defining the bounding box more precisely. If you made a mistake, click "Reset" button to start over. "Show VOI" button can be used to show/hide blue overlay of the region of interest of the the image slice views. |
- Step 3: Select tumor segmentation threshold
| Use threshold control slider to find the intensity that most closely approximates tumor volume. Tumor volume is rendered in the 3D viewer as you are choosing the appropriate threshold, and is also visualized as semi-transparent label in the image slice viewers. |
- Step 4: Define metric
| We provide two metrics -- intensity-based and deformation-based -- to quantify tumor progression (see details in this paper). Choose the metric you would like to use. |
- Analysis/Results
| Results are reported as the change in tumor volume, separately for growth and shrinkage component. The quantitative results are reported in voxels and mm^3.
Sensitivity of the analysis can be controlled by choosing Sensitive, Moderate, or Robust analysis approach (Robust being the most conservative). As you change the sensitivity, visualization and quantitative data are updated accordingly. Growth is shown in purple, and shrinkage -- in green color. You can choose to display the original sampling grid, and view the coronal slice of the subject using the module controls. |
Development
Dependencies
| ChangeTracker is using Rigid Registration module through the CommandLineModule shared object invocation. Note, that there are two distinct rigid registration modules in Slicer, which have identical functionality: "Linear registration" and "Rigid registration". Both of these modules should be available in your build, but "Rigid registration" *must* be available for ChangeTracker to function properly. You can verify the availability of the module manually by looking at the list of modules in the registration category. |
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Follow this link to the ChangeTracker source code in ViewVC.
Documentation generated by doxygen.
More Information
Acknowledgment
ChangeTracker development has been funded by Brain Science Foundation
References
- E.Konukoglu, W.M.Wells, S.Novellas, N.Ayache, R.Kikinis, P.M.Black, K.M.Pohl. Monitoring Slowly Evolving Tumors. Proc. of 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro 2008, pp.812-815 link