Difference between revisions of "Slicer 4.0:Training"
From Slicer Wiki
m (Text replacement - "\[http:\/\/www\.slicer\.org\/slicerWiki\/images\/([^ ]+) ([^]]+)]" to "[http://www.slicer.org/w/img_auth.php/$1 $2]") |
|||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 18: | Line 18: | ||
| id="Slicer3MinuteTutorial" style="background:#CCFF99; color:blue; font-size:110%" align="center"| '''Core tutorial 1''' | | id="Slicer3MinuteTutorial" style="background:#CCFF99; color:blue; font-size:110%" align="center"| '''Core tutorial 1''' | ||
| style="background:#CCFF99; color:black" valign="top"| '''[[Media:Slicer3Minute_SoniaPujol_3.6.1.pdf | Slicer3Minute Tutorial]]'''<br><br><br>1)open slicer. 2)opened data set. 3)rotated model. 4)clicked slice visibility icon (red). 5)turn skin opacity to 0.0. 6) clicked slice visibility icon (green). 7)clip the scull and bone exposing the white matter tissue. 8) move through the scan (green). 9)take visibility off white matter. 10) zoom in and out. | | style="background:#CCFF99; color:black" valign="top"| '''[[Media:Slicer3Minute_SoniaPujol_3.6.1.pdf | Slicer3Minute Tutorial]]'''<br><br><br>1)open slicer. 2)opened data set. 3)rotated model. 4)clicked slice visibility icon (red). 5)turn skin opacity to 0.0. 6) clicked slice visibility icon (green). 7)clip the scull and bone exposing the white matter tissue. 8) move through the scan (green). 9)take visibility off white matter. 10) zoom in and out. | ||
| − | | style="background:#CCFF99; color:black" valign="top" | '''[http://www.slicer.org/ | + | | style="background:#CCFF99; color:black" valign="top" | '''[http://www.slicer.org/w/img_auth.php/5/51/Slicer3MinuteDataset.zip Slicer3Minute Data]'''<br><br> step 2) picture for tutorial is out of date. |
| style="background:#CCFF99; color:black" align="center"| [[Image:Slicer3Minute_3.6RC2.png |250px]] | | style="background:#CCFF99; color:black" align="center"| [[Image:Slicer3Minute_3.6RC2.png |250px]] | ||
|- | |- | ||
| id="Slicer3VisualizationTutorial" style="background:#FFCC99; color:blue; font-size:110%" align="center"| '''Core tutorial 2''' | | id="Slicer3VisualizationTutorial" style="background:#FFCC99; color:blue; font-size:110%" align="center"| '''Core tutorial 2''' | ||
| style="background:#FFCC99; color:black" valign="top"| '''[[media:Slicer3_DataLoadingAndVisualization_UCSF2010_SoniaPujol.pdf | Slicer3Visualization Tutorial]]'''<br><br><br>1) open slicer. 2) parts of slicer. 3) load volume 001.dcm from dicom directory. 4) open module volumes. 5) using window/level slider. 6) open volume spgr.nhdr. 7) select red only layout from the choose view icon. 8) fit the image size to window. 9) click foreground icon and select the dicom image. 10)change display using the background/foreground icons. 11)use slider to display ventricles. 12) go back to standard view. PART 2 1) select volume file.<br><br> | | style="background:#FFCC99; color:black" valign="top"| '''[[media:Slicer3_DataLoadingAndVisualization_UCSF2010_SoniaPujol.pdf | Slicer3Visualization Tutorial]]'''<br><br><br>1) open slicer. 2) parts of slicer. 3) load volume 001.dcm from dicom directory. 4) open module volumes. 5) using window/level slider. 6) open volume spgr.nhdr. 7) select red only layout from the choose view icon. 8) fit the image size to window. 9) click foreground icon and select the dicom image. 10)change display using the background/foreground icons. 11)use slider to display ventricles. 12) go back to standard view. PART 2 1) select volume file.<br><br> | ||
| − | | style="background:#FFCC99; color:black" valign="top" | '''[http://www.slicer.org/ | + | | style="background:#FFCC99; color:black" valign="top" | '''[http://www.slicer.org/w/img_auth.php/6/61/Slicer3VisualizationDataset.zip Slicer3Visualization Data]'''<br><br>step 2)edit tutorial no progress bar. step 3)a)slicer does not display the dicom header information, b) new tutorial image is needed c)click open not apply. step 5) new tutorial image is needed. step 11)scans improperly under (-30) (red) unsure if it is slicer or the selected data set. PART 2 step 1)the loading menu is missing, unable to do any more on this tutorial without it. |
| style="background:#FFCC99; color:black" align="center"| [[Image:VisualizationTutorial 3.6RC3.png|250px]] | | style="background:#FFCC99; color:black" align="center"| [[Image:VisualizationTutorial 3.6RC3.png|250px]] | ||
|- | |- | ||
| Line 38: | Line 38: | ||
| id="InteractiveEditor" style="background:#FFCC99; color:blue; font-size:110%" align="center"| '''Core tutorial 5''' | | id="InteractiveEditor" style="background:#FFCC99; color:blue; font-size:110%" align="center"| '''Core tutorial 5''' | ||
| style="background:#FFCC99; color:black" valign="top"|'''[[media:InteractiveEditorTutorial_Slicer3.6-SoniaPujol.pdf |Interactive Editor]] '''<br><br><br>1) open slicer. 2) upload "grayscale.nrrd". 3) open editor module. 4) set master volume to grayscale. 5) select "GenericAtonomyColors" Color map and apply it. <br>'''Audience:''' All users and developers. | | style="background:#FFCC99; color:black" valign="top"|'''[[media:InteractiveEditorTutorial_Slicer3.6-SoniaPujol.pdf |Interactive Editor]] '''<br><br><br>1) open slicer. 2) upload "grayscale.nrrd". 3) open editor module. 4) set master volume to grayscale. 5) select "GenericAtonomyColors" Color map and apply it. <br>'''Audience:''' All users and developers. | ||
| − | | style="background:#FFCC99; color:black" valign="top"|'''[ | + | | style="background:#FFCC99; color:black" valign="top"|'''[[File:EditorTutorialDataset.zip|Editor Data]]'''<br><br>step 5) the editor does not display the frame that contains different tools for editing. |
| style="background:#FFCC99; color:black" align="center"|[[Image:InteractiveEditor.png|250px]] | | style="background:#FFCC99; color:black" align="center"|[[Image:InteractiveEditor.png|250px]] | ||
|- | |- | ||
| id="ManualRegistration" style="background:#FFCC33; color:blue; font-size:110%" align="center"| '''Core tutorial 6''' | | id="ManualRegistration" style="background:#FFCC33; color:blue; font-size:110%" align="center"| '''Core tutorial 6''' | ||
| style="background:#FFCC33; color:black" valign="top"|'''[[media:ManualRegistration_Slicer3.6.pdf | Manual Registration]] ''' <br><br><br> 1) open slicer. 2)upload "ManualRegistrationExample_SlicerScene.mrml". 3)select 4-up. 4) link views. 5) set foreground to moving. 6)set background to fixed. 7)set the visibility slider to halfway between the two. 8) open data module. 9) select the new node. 10)rename it Manuallnit. 11) drag MRI_Moving on top of Manuallnit. 12) open the transforms module. 13) Move slider "IS" to (-21). 14) move slider "PA" to (-24). 15) move slider "IS" to 14. 16) move slider "LR" to 26. 17)save file! <br><br>'''Audience:''' First time & early users. | | style="background:#FFCC33; color:black" valign="top"|'''[[media:ManualRegistration_Slicer3.6.pdf | Manual Registration]] ''' <br><br><br> 1) open slicer. 2)upload "ManualRegistrationExample_SlicerScene.mrml". 3)select 4-up. 4) link views. 5) set foreground to moving. 6)set background to fixed. 7)set the visibility slider to halfway between the two. 8) open data module. 9) select the new node. 10)rename it Manuallnit. 11) drag MRI_Moving on top of Manuallnit. 12) open the transforms module. 13) Move slider "IS" to (-21). 14) move slider "PA" to (-24). 15) move slider "IS" to 14. 16) move slider "LR" to 26. 17)save file! <br><br>'''Audience:''' First time & early users. | ||
| − | | style="background:#FFCC33; color:black" valign="top"|'''[http://www.slicer.org/ | + | | style="background:#FFCC33; color:black" valign="top"|'''[http://www.slicer.org/w/img_auth.php/8/88/Slicer3_Tutorial_ManualRegistration_ExampleDataset.zip Manual Registration Data]'''<br><br>step 10) tutorial says to enter it in a name field but on Slicer4 you need to double-click it. step 13) you must first select transform node: manuallnit. |
| style="background:#FFCC33; color:black" align="center"|[[Image:Slicer3_ManualRegistrationTutorial.gif|250px]] | | style="background:#FFCC33; color:black" align="center"|[[Image:Slicer3_ManualRegistrationTutorial.gif|250px]] | ||
|- | |- | ||
| id="DiffusionMRITutorial" style="background:#FFCC99; color:blue; font-size:110%" align="center"| '''Specialized tutorial 1''' | | id="DiffusionMRITutorial" style="background:#FFCC99; color:blue; font-size:110%" align="center"| '''Specialized tutorial 1''' | ||
| style="background:#FFCC99; color:black" valign="top"|'''[[Media:DiffusionMRITutorial_UCSF2010_SoniaPujol.pdf | Diffusion MRI tutorial]]''' <br><br><br>1) open slicer. 2) upload dwiDataset.nhdr. 3) open Volumes module. 4) set dwiDataset as Active Volume. 5) open the Diffusion Tensor Estimation module. 6) plug in the dwiDataset as the Input DWI Volume. 7) set output to create new DiffusionTensorVolume. 8) set Otsu Threshold Mask to Create New Volume. 9) select tensor Estimation Algorithm LS. 10) click apply. <br><br>'''Audience:''' All users and developers. | | style="background:#FFCC99; color:black" valign="top"|'''[[Media:DiffusionMRITutorial_UCSF2010_SoniaPujol.pdf | Diffusion MRI tutorial]]''' <br><br><br>1) open slicer. 2) upload dwiDataset.nhdr. 3) open Volumes module. 4) set dwiDataset as Active Volume. 5) open the Diffusion Tensor Estimation module. 6) plug in the dwiDataset as the Input DWI Volume. 7) set output to create new DiffusionTensorVolume. 8) set Otsu Threshold Mask to Create New Volume. 9) select tensor Estimation Algorithm LS. 10) click apply. <br><br>'''Audience:''' All users and developers. | ||
| − | | style="background:#FFCC99; color:black" valign="top" |'''[http://www.slicer.org/ | + | | style="background:#FFCC99; color:black" valign="top" |'''[http://www.slicer.org/w/img_auth.php/c/cf/DiffusionDataset.zip Diffusion Data]''' step 8)there is no setting called Otsu Threshold Mask, i tried twice once replacing Otsu Mask with output baseline volume, because Diffusion Tensor Mask has no Create New Volume setting. 10) project can go no further due to errors in step 8. |
| style="background:#FFCC99; color:black" align="center"|[[Image:cc.PNG |250px]] | | style="background:#FFCC99; color:black" align="center"|[[Image:cc.PNG |250px]] | ||
|- | |- | ||
| − | | id="ChangeTrackerTutorial" style="background:# | + | | id="ChangeTrackerTutorial" style="background:#FFCC99; color:blue; font-size:110%" align="center"| '''Specialized tutorial 2''' |
| − | | style="background:# | + | | style="background:#FFCC99; color:black" valign="top"|'''[[media:Slicer3.6-ChangeTrackerTutorial.pdf | Change Tracker Tutorial]]''' <br><br><br>1)open slicer 2) go to modules then Wizards then Change Tracker <br><br>'''Audience:''' All users interested in longitudinal analysis of pathology. |
| − | | style="background:# | + | | style="background:#FFCC99; color:black" valign="top" |step 2) the Wizards menu option does not expand, nor does IGT, but all the others work fine. |
| − | | style="background:# | + | | style="background:#FFCC99; color:black" align="center"|[[Image:Slicer3.4.1-ChangeTracker.jpg|250px]] |
|- | |- | ||
| − | | id="FreeSurferCourse" style="background:# | + | | id="FreeSurferCourse" style="background:#FFCC99; color:blue; font-size:110%" align="center"| '''Specialized tutorial 3''' |
| − | | style="background:# | + | | style="background:#FFCC99; color:black" valign="top"|'''[[media:FreeSurferCourse_Slicer3.6_SoniaPujol.pdf | FreeSurfer Course ]]'''<br><br> 1) open slicer. 2) go to Volumes module. 3) click on select volume file. 4) open brain.mgz centered. 5)link the slices and make them visible. 6) repeat step 3. 7) click label map and apply. 8) click look up table than FreeSurferLables. 9)look through labels. <br> PART 2 <br> 1) select model maker from modules. 2) make input volume aseg. 3) select models make a new hierarchy. 4)name the model right-hippocampus and uncheck generate all models. 5)under parameters enter #53 as a label. 6)turn off slice visibility. 7) apply. |
<br><br>'''Audience:''' All users | <br><br>'''Audience:''' All users | ||
| − | | style="background:# | + | | style="background:#FFCC99; color:black" valign="top" |'''[http://www.na-mic.org/Wiki/index.php/File:FreeSurferTutorialData.zip FreeSurfer tutorial data ''']<br><br> step 3)does not exist instead i went through the file menu. step 6) same problem as step 3, same solution. step 7) instead click volume information then click label map. <br>PART 2<br>step 7) "completed with errors" no image is displayed. |
| − | | style="background:# | + | | style="background:#FFCC99; color:black" align="center|[[Image:FreeSurferTutorial.PNG|250px]] |
|- | |- | ||
| id="NeurosurgicalPlanningTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | | id="NeurosurgicalPlanningTutorial" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| Line 74: | Line 74: | ||
| id="EMSegmenterSimpleMode" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | | id="EMSegmenterSimpleMode" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| style="background:#FFFFCC; color:black" valign="top"| [[media:EMSegmenterTutorialSimpleMode-Slicer-3.6.3-1.pdf | '''EMSegmenter Simple Mode ''']] <br><br> This tutorial takes the trainee through the segmentation of a MRI Human Brain without to adjust any parameters. | | style="background:#FFFFCC; color:black" valign="top"| [[media:EMSegmenterTutorialSimpleMode-Slicer-3.6.3-1.pdf | '''EMSegmenter Simple Mode ''']] <br><br> This tutorial takes the trainee through the segmentation of a MRI Human Brain without to adjust any parameters. | ||
| − | | style="background:#FFFFCC; color:black" valign="top" | [http://www.slicer.org/ | + | | style="background:#FFFFCC; color:black" valign="top" | [http://www.slicer.org/w/img_auth.php/c/cd/MRIHumanBrain_T1_aligned.nrrd '''Human Brain T1 Data'''] |
| style="background:#FFFFCC; color:black" align="center"| [[Image:EMSegmenter-Simple-Mode.png|250px]] | | style="background:#FFFFCC; color:black" align="center"| [[Image:EMSegmenter-Simple-Mode.png|250px]] | ||
|- | |- | ||
| id="EMSegmenterAdvancedMode" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | | id="EMSegmenterAdvancedMode" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| style="background:#FFFFCC; color:black" valign="top"| [[media:EMSegmenterTutorialAdvancedMode-Slicer-3.6.3-2.pdf | '''EMSegmenter Advanced Mode ''']] <br><br> This tutorial takes the trainee through the segmentation of a MRI Human Brain. The trainee will learn how to setup the EMSegmenter, this includes the creation of a task, the creation of an anatomical tree and adjusting the weights for the EM algorithm. | | style="background:#FFFFCC; color:black" valign="top"| [[media:EMSegmenterTutorialAdvancedMode-Slicer-3.6.3-2.pdf | '''EMSegmenter Advanced Mode ''']] <br><br> This tutorial takes the trainee through the segmentation of a MRI Human Brain. The trainee will learn how to setup the EMSegmenter, this includes the creation of a task, the creation of an anatomical tree and adjusting the weights for the EM algorithm. | ||
| − | | style="background:#FFFFCC; color:black" valign="top" | [http://www.slicer.org/ | + | | style="background:#FFFFCC; color:black" valign="top" | [http://www.slicer.org/w/img_auth.php/c/cd/MRIHumanBrain_T1_aligned.nrrd '''Human Brain T1 Data'''] |
| style="background:#FFFFCC; color:black" align="center"| [[Image:EMSegmenter-Advanced-Mode.png|250px]] | | style="background:#FFFFCC; color:black" align="center"| [[Image:EMSegmenter-Advanced-Mode.png|250px]] | ||
|- | |- | ||
| id="OpenIGTLink" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | | id="OpenIGTLink" style="background:#FFFFCC; color:blue; font-size:110%" align="center"| '''Specialized''' | ||
| style="background:#FFFFCC; color:black" valign="top"|[[media:OpenIGTLinkTutorial_Slicer3.6.2_JunichiTokuda_Dec2010.pdf | '''OpenIGTLink''']] <br><br> This tutorial shows to connect an IGT device using the OpenIGTLink. | | style="background:#FFFFCC; color:black" valign="top"|[[media:OpenIGTLinkTutorial_Slicer3.6.2_JunichiTokuda_Dec2010.pdf | '''OpenIGTLink''']] <br><br> This tutorial shows to connect an IGT device using the OpenIGTLink. | ||
| − | |style="background:#FFFFCC; color:black" valign="top" | [ | + | |style="background:#FFFFCC; color:black" valign="top" | [[Modules:OpenIGTLinkIF-3.6-Simulators'''OpenIGTLink|Data Simulator''']] |
| style="background:#FFFFCC; color:black" align="center"| [[Image:OpenIGT.PNG|250px]] | | style="background:#FFFFCC; color:black" align="center"| [[Image:OpenIGT.PNG|250px]] | ||
|- | |- | ||
Latest revision as of 12:39, 27 November 2019
Home < Slicer 4.0:TrainingContents
Slicer 4.0 Tutorials
Color chart
green - perfect
yellow - problems but nothing too serious
red - i was unable to finish the tutorial due to problems
| Category | Steps Taken | Problems | Image |
| Core tutorial 1 | Slicer3Minute Tutorial 1)open slicer. 2)opened data set. 3)rotated model. 4)clicked slice visibility icon (red). 5)turn skin opacity to 0.0. 6) clicked slice visibility icon (green). 7)clip the scull and bone exposing the white matter tissue. 8) move through the scan (green). 9)take visibility off white matter. 10) zoom in and out. |
Slicer3Minute Data step 2) picture for tutorial is out of date. |
|
| Core tutorial 2 | Slicer3Visualization Tutorial 1) open slicer. 2) parts of slicer. 3) load volume 001.dcm from dicom directory. 4) open module volumes. 5) using window/level slider. 6) open volume spgr.nhdr. 7) select red only layout from the choose view icon. 8) fit the image size to window. 9) click foreground icon and select the dicom image. 10)change display using the background/foreground icons. 11)use slider to display ventricles. 12) go back to standard view. PART 2 1) select volume file. |
Slicer3Visualization Data step 2)edit tutorial no progress bar. step 3)a)slicer does not display the dicom header information, b) new tutorial image is needed c)click open not apply. step 5) new tutorial image is needed. step 11)scans improperly under (-30) (red) unsure if it is slicer or the selected data set. PART 2 step 1)the loading menu is missing, unable to do any more on this tutorial without it. |
|
| skip | Programming in Slicer3 Tutorial 1)open file "helloworld.xml" and "helloworld.cxx" and "Cmakelists.txt". 2)edit the file "Cmakelists.txt". 3)open the Cmake executable. Audience: Programmers and Engineers. |
HelloWorld Plugin step 3) the tutorial shows the wrong location for the Cmake executable, it says "Slicer3-lib/CMake-build/bin" so i tried going through Slicer-4-libs but there is no CMake or build option. |
|
| skip | Hello Python Tutorial The Hello Python tutorial is an introduction to the integration of Python stand-alone programs outside of the Slicer3 source tree. Audience: Programmers and Engineers. |
HelloPython Plugin The HelloPython tutorial dataset contains an MR scan of the brain and pre-computed xml and Python code for integrating the Hello Python plug-in to Slicer3. |
|
| Core tutorial 5 | Interactive Editor 1) open slicer. 2) upload "grayscale.nrrd". 3) open editor module. 4) set master volume to grayscale. 5) select "GenericAtonomyColors" Color map and apply it. Audience: All users and developers. |
File:EditorTutorialDataset.zip step 5) the editor does not display the frame that contains different tools for editing. |
|
| Core tutorial 6 | Manual Registration 1) open slicer. 2)upload "ManualRegistrationExample_SlicerScene.mrml". 3)select 4-up. 4) link views. 5) set foreground to moving. 6)set background to fixed. 7)set the visibility slider to halfway between the two. 8) open data module. 9) select the new node. 10)rename it Manuallnit. 11) drag MRI_Moving on top of Manuallnit. 12) open the transforms module. 13) Move slider "IS" to (-21). 14) move slider "PA" to (-24). 15) move slider "IS" to 14. 16) move slider "LR" to 26. 17)save file! Audience: First time & early users. |
Manual Registration Data step 10) tutorial says to enter it in a name field but on Slicer4 you need to double-click it. step 13) you must first select transform node: manuallnit. |
|
| Specialized tutorial 1 | Diffusion MRI tutorial 1) open slicer. 2) upload dwiDataset.nhdr. 3) open Volumes module. 4) set dwiDataset as Active Volume. 5) open the Diffusion Tensor Estimation module. 6) plug in the dwiDataset as the Input DWI Volume. 7) set output to create new DiffusionTensorVolume. 8) set Otsu Threshold Mask to Create New Volume. 9) select tensor Estimation Algorithm LS. 10) click apply. Audience: All users and developers. |
Diffusion Data step 8)there is no setting called Otsu Threshold Mask, i tried twice once replacing Otsu Mask with output baseline volume, because Diffusion Tensor Mask has no Create New Volume setting. 10) project can go no further due to errors in step 8. | 
|
| Specialized tutorial 2 | Change Tracker Tutorial 1)open slicer 2) go to modules then Wizards then Change Tracker Audience: All users interested in longitudinal analysis of pathology. |
step 2) the Wizards menu option does not expand, nor does IGT, but all the others work fine. | |
| Specialized tutorial 3 | FreeSurfer Course 1) open slicer. 2) go to Volumes module. 3) click on select volume file. 4) open brain.mgz centered. 5)link the slices and make them visible. 6) repeat step 3. 7) click label map and apply. 8) click look up table than FreeSurferLables. 9)look through labels. PART 2 1) select model maker from modules. 2) make input volume aseg. 3) select models make a new hierarchy. 4)name the model right-hippocampus and uncheck generate all models. 5)under parameters enter #53 as a label. 6)turn off slice visibility. 7) apply.
|
FreeSurfer tutorial data step 3)does not exist instead i went through the file menu. step 6) same problem as step 3, same solution. step 7) instead click volume information then click label map. PART 2 step 7) "completed with errors" no image is displayed. |
|
| Specialized | Neurosurgical Planning Tutorial This tutorial takes the trainee through a complete workup for neurosurgical patient-specific mapping. Also see this tutorial for information on how to use Slicer's affine registration, simple region growing, model maker and tractography modules. Audience: All users interested in image-guided therapy. |
Neurosurgical Planning Data | 
|
| Specialized | PET/CT SUV Tutorial This tutorial takes the trainee through the computation of SUV body weight on a baseline and followup study. |
PET/CT Data | 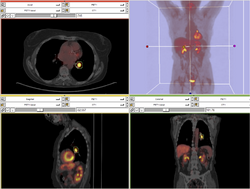
|
| Specialized | EMSegmenter Simple Mode This tutorial takes the trainee through the segmentation of a MRI Human Brain without to adjust any parameters. |
Human Brain T1 Data | 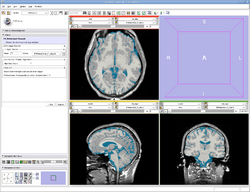
|
| Specialized | EMSegmenter Advanced Mode This tutorial takes the trainee through the segmentation of a MRI Human Brain. The trainee will learn how to setup the EMSegmenter, this includes the creation of a task, the creation of an anatomical tree and adjusting the weights for the EM algorithm. |
Human Brain T1 Data | 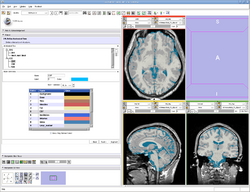
|
| Specialized | OpenIGTLink This tutorial shows to connect an IGT device using the OpenIGTLink. |
Data Simulator | 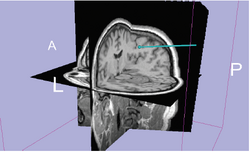
|
| Workflow | White Matter Exploration for Neurosurgical Planning This tutorial walks the user through a workflow for exploring white matter fibers surrounding a tumor using Diffusion Tensor Imaging (DTI) Tractography. |
White Matter Exploration dataset | 
|