Difference between revisions of "Documentation/4.2/Modules/MultiVolumeExplorer"
m (Text replacement - "http://wiki.slicer.org/slicerWiki/images/" to "http://www.slicer.org/w/img_auth.php/") |
|||
| (5 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| + | <noinclude>{{documentation/versioncheck}}</noinclude> | ||
{{ambox | {{ambox | ||
| type = protection | | type = protection | ||
| Line 41: | Line 42: | ||
Most frequently used for these scenarios: | Most frequently used for these scenarios: | ||
* visualization of a DICOM dataset that contains multiple frames that can be separated based on some tag (e.g., DCE MRI data, where individual temporally resolved frames are identified by Trigger Time tag (0018,1060) | * visualization of a DICOM dataset that contains multiple frames that can be separated based on some tag (e.g., DCE MRI data, where individual temporally resolved frames are identified by Trigger Time tag (0018,1060) | ||
| − | * visualization of multiple frames defined in the same coordinate frame, saved as individual volumes in NRRD, NIfTI, or any other [[Documentation/ | + | * visualization of multiple frames defined in the same coordinate frame, saved as individual volumes in NRRD, NIfTI, or any other [[Documentation/{{documentation/version}}/SlicerApplication/SupportedDataFormat|image format supported by 3D Slicer]] |
* exploration of the multivolume data (cine mode visualization, plotting, volume rendering) | * exploration of the multivolume data (cine mode visualization, plotting, volume rendering) | ||
| Line 50: | Line 51: | ||
* [http://www.youtube.com/watch?v=7zFWRzC31o4 Screencast tutorial] (note that the module is located in '''Modules -> Work in progress -> MultiVolume Support''', not in Modules -> MultiVolume Support, as shown in the video) | * [http://www.youtube.com/watch?v=7zFWRzC31o4 Screencast tutorial] (note that the module is located in '''Modules -> Work in progress -> MultiVolume Support''', not in Modules -> MultiVolume Support, as shown in the video) | ||
* Sample datasets are available: | * Sample datasets are available: | ||
| − | ** [http:// | + | ** Anonymized prostate DCE MRI DICOM series: |
| + | *** [http://www.slicer.org/w/img_auth.php/5/56/Case1_DCE.tgz tgz version] | ||
| + | *** [http://www.slicer.org/w/img_auth.php/c/c2/DCE_series.zip zip version] | ||
** [[File:Cardiac_ECGg_CT.tgz]] (ECG-gated contrast-enhanced cardiac CT) (use "User-defined non-DICOM" parameter option to load it into MultiVolumeImporter). Short [[media:BeatingHeart.mov|movie]] | ** [[File:Cardiac_ECGg_CT.tgz]] (ECG-gated contrast-enhanced cardiac CT) (use "User-defined non-DICOM" parameter option to load it into MultiVolumeImporter). Short [[media:BeatingHeart.mov|movie]] | ||
| Line 71: | Line 74: | ||
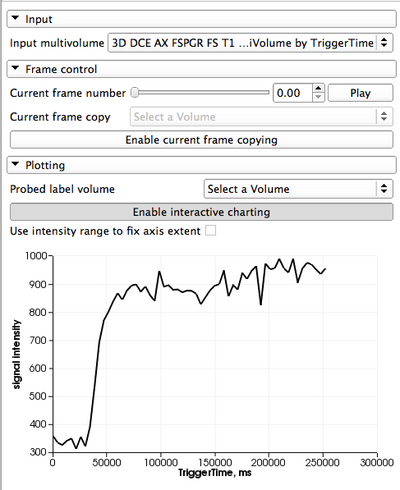
** ''Static plotting'': This mode of plotting is enabled by the [[Documentation/4.2/SlicerApplication/MainApplicationGUI#Chart_Viewers | Charts]] infrastructure of Slicer, which is exposed in any of the ''Quantitive layouts''. To use this mode, follow these steps: | ** ''Static plotting'': This mode of plotting is enabled by the [[Documentation/4.2/SlicerApplication/MainApplicationGUI#Chart_Viewers | Charts]] infrastructure of Slicer, which is exposed in any of the ''Quantitive layouts''. To use this mode, follow these steps: | ||
**# Extract a representative frame from your multivolume dataset, as discussed above. | **# Extract a representative frame from your multivolume dataset, as discussed above. | ||
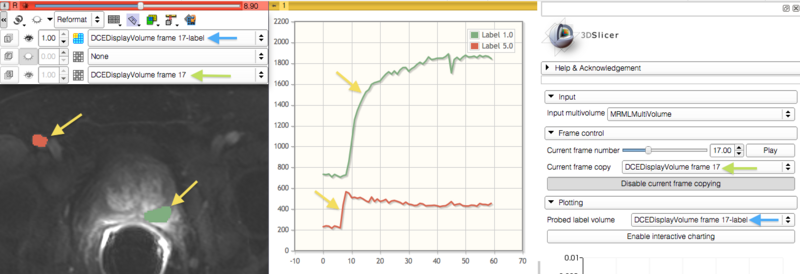
| − | **# Use the [[Documentation/ | + | **# Use the [[Documentation/{{documentation/version}}/Modules/Editor|Editor]] module to create labels for the regions of interest. |
**# Select the prepared label volume in the ''Probed label volume'' selector in the ''Plotting'' panel. The ''Charts'' viewer in the Slicer Quantitative layout will be updated with the average intensity values for each of the contoured ROIs. The colors of the plots will be the same as the colors of ROI labels. | **# Select the prepared label volume in the ''Probed label volume'' selector in the ''Plotting'' panel. The ''Charts'' viewer in the Slicer Quantitative layout will be updated with the average intensity values for each of the contoured ROIs. The colors of the plots will be the same as the colors of ROI labels. | ||
| | | | ||
| Line 88: | Line 91: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Related Modules}} | {{documentation/{{documentation/version}}/module-section|Related Modules}} | ||
| − | Before multivolume data can be viewed/explored, it has to be loaded using either [[Documentation/ | + | Before multivolume data can be viewed/explored, it has to be loaded using either [[Documentation/{{documentation/version}}/Modules/DICOM|DICOM]] (if the dataset is in DICOM format) or [[Documentation/{{documentation/version}}/Modules/MultiVolumeImporter|MultVolumeImporter]] modules. |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 12:31, 27 November 2019
Home < Documentation < 4.2 < Modules < MultiVolumeExplorer
|
For the latest Slicer documentation, visit the read-the-docs. |
WARNING: This module is Work in Progress, which means:
|
Introduction and Acknowledgements
|
This work is supported by NA-MIC, NAC, NCIGT, and the Slicer Community. This work is partially supported by the following grants: P41EB015898, P41RR019703, R01CA111288 and U01CA151261. | |||||||||||
|
Module Description
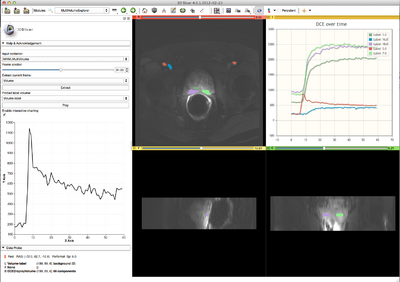
This module provides support for exploring multivolume (multiframe) data.
Use Cases
Most frequently used for these scenarios:
- visualization of a DICOM dataset that contains multiple frames that can be separated based on some tag (e.g., DCE MRI data, where individual temporally resolved frames are identified by Trigger Time tag (0018,1060)
- visualization of multiple frames defined in the same coordinate frame, saved as individual volumes in NRRD, NIfTI, or any other image format supported by 3D Slicer
- exploration of the multivolume data (cine mode visualization, plotting, volume rendering)
Tutorials
- Screencast tutorial (note that the module is located in Modules -> Work in progress -> MultiVolume Support, not in Modules -> MultiVolume Support, as shown in the video)
- Sample datasets are available:
- Anonymized prostate DCE MRI DICOM series:
- File:Cardiac ECGg CT.tgz (ECG-gated contrast-enhanced cardiac CT) (use "User-defined non-DICOM" parameter option to load it into MultiVolumeImporter). Short movie
Panels and their use
MultiVolume data is represented in this module using two components:
- MultiVolume node: this is the node you select when you import the dataset using MultiVolumeImporter module
- DisplayNode: this is the node responsible for visualization and storage of the actual image data. The name of that latter node is assigned based on the type of data you are importing. For example, if you import "4D DCE" dataset, it will be called "DCEDisplayNode". This is the node you need to choose in slice viewers to see the multivolume dataset.
|
Related Modules
Before multivolume data can be viewed/explored, it has to be loaded using either DICOM (if the dataset is in DICOM format) or MultVolumeImporter modules.
References
- Development of this module was initiated at the 2012 NA-MIC Project week at SLC (see http://wiki.na-mic.org/Wiki/index.php/2012_Project_Week:4DImageSlicer4)
Information for Developers
This module is an external Slicer module. The source code is available on Github here: https://github.com/fedorov/MultiVolumeExplorer
Features to be implemented
- allow the user to speed-up or slow-down the playback, and also turn off "loop"
- report to the user the actual, measured, frame rate
Features under consideration
- can we use chart viewer for interactive plotting? -- Discussed with Jim, speedup is possible, but not a priority right now
- interactive update of the active frame based on the point selected in the chart viewer
- add GUI elements to update the current frame at the exact time intervals specified in multivolume. If the intervals are non-uniform, I am not sure I can use the timer anymore, so this would require some extra thought.
- import multivolumes from ITK 4d images