Difference between revisions of "Documentation/Nightly/Modules/GrowCutSegmentation"
Hillarylia (talk | contribs) (Began new GrowCut effect page to be linked into editor module page) |
m (Text replacement - "\[https:\/\/www\.slicer\.org\/slicerWiki\/index.php\/([^ ]+) ([^]]+)]" to "$2") |
||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 5: | Line 5: | ||
{{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | {{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | ||
| + | This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from [http://www.ncbcs.org/ National Centers for Biomedical Computing]. | ||
| + | |||
Authors: Harini Veeraraghavan, Jim Miller | Authors: Harini Veeraraghavan, Jim Miller | ||
| − | + | ||
| − | + | Contact: veerarag@ge.com | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 13: | Line 15: | ||
{| | {| | ||
| | | | ||
| − | Grow Cut Segmentation is a competitive region growing algorithm using cellular automata. The algorithm works by using a set of user input scribbles for foreground and background. For N-class segmentation, the algorithm requires a set of scribbles corresponding the N classes and a scribble for a don't care class. The algorithm executes as follows: | + | Grow Cut Segmentation is a competitive region growing algorithm using cellular automata found as an effect in the [[Documentation/Nightly/Modules/Editor|Editor module]]. The algorithm works by using a set of user input scribbles for the foreground and background. For N-class segmentation, the algorithm requires a set of scribbles corresponding the N classes and a scribble for a don't care class. |
| − | Using the "user input scribbles", the algorithm automatically computes a region of interest that encompass the scribbles. It is important that the scribbles do not include a single voxel out of your region of interest as the | + | |
| − | Next, the algorithm iteratively tries to label all the pixels in the image using the label of pixels in the user scribbled portions of the image. | + | The algorithm executes as follows: |
| − | The algorithm converges when all the pixels in the ROI are labeled, and no pixel can change it's label any more. | + | Using the "user input scribbles", the algorithm automatically computes a region of interest that encompass the scribbles. ''It is important that the scribbles do not include a single voxel out of your region of interest as the seeding is sensitive and will not respond well to outliers.'' Next, the algorithm iteratively tries to label all the pixels in the image using the label of pixels in the user scribbled portions of the image. The algorithm converges when all the pixels in the ROI are labeled, and no pixel can change it's label any more. Individual pixels are labeled by computing a weighted similarity metric of a pixel with all its neighbors, where the weights correspond to the neighboring pixel's strength. The neighbor that results in the largest weight greater than the given pixel's strength, confers its label to the given pixel. After the segmentation, the user can edit the segmentation by providing additional gestures in the image as illustrated in the figure below. |
| − | Individual pixels are labeled by computing a weighted similarity metric of a pixel with all its neighbors, where the weights correspond to the neighboring pixel's strength. The neighbor that results in the largest weight greater than the given pixel's strength, confers its label to the given pixel. | ||
| − | After the segmentation, the user can edit the segmentation by providing additional gestures in the image as illustrated in the figure below. | ||
| | | | ||
|} | |} | ||
| + | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|Screen Captures}} | ||
| + | |||
| + | |||
| + | <gallery widths=250px heights=250px class="center"> | ||
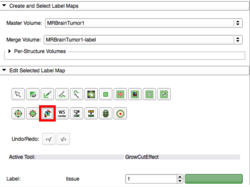
| + | File:GrowCutInputPanel-2016-06-14.png|Input Panel, select the GrowCut icon | ||
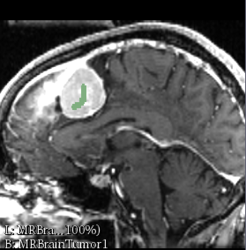
| + | File:GrowCutScribblePos-2016-06-15.png|Scribble Positive Gestures | ||
| + | File:GrowCutScribbleNeg-2016-06-15.png|Scribble Negative Gestures with a different label value | ||
| + | File:GrowCutResult-2016-06-15.png|Resulting segmentation. This can be altered by providing additional gestures. | ||
| + | </gallery> | ||
| + | |||
| + | ''Click to enlarge photos if necessary'' | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| − | {{documentation/{{documentation/version}}/module-section|Similar | + | {{documentation/{{documentation/version}}/module-section|Similar Effects}} |
| + | See also the [[Documentation/4.3/Modules/FastGrowCut|Fast GrowCut]] documentation. This is a fast version of the GrowCut algorithm. It is downloadable in the Extension Manager, thus requires memory. | ||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|References}} | ||
| + | *V. Vezhnevets and V. Konouchine, "GrowCut - Interactive multi-label N-D image segmentation", in Proc. Graphicon, 2005. pp. 150--156. | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-footer}} | {{documentation/{{documentation/version}}/module-footer}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 02:40, 27 November 2019
Home < Documentation < Nightly < Modules < GrowCutSegmentation
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from National Centers for Biomedical Computing.
Authors: Harini Veeraraghavan, Jim Miller
Contact: veerarag@ge.com
Module Description
|
Grow Cut Segmentation is a competitive region growing algorithm using cellular automata found as an effect in the Editor module. The algorithm works by using a set of user input scribbles for the foreground and background. For N-class segmentation, the algorithm requires a set of scribbles corresponding the N classes and a scribble for a don't care class. The algorithm executes as follows: Using the "user input scribbles", the algorithm automatically computes a region of interest that encompass the scribbles. It is important that the scribbles do not include a single voxel out of your region of interest as the seeding is sensitive and will not respond well to outliers. Next, the algorithm iteratively tries to label all the pixels in the image using the label of pixels in the user scribbled portions of the image. The algorithm converges when all the pixels in the ROI are labeled, and no pixel can change it's label any more. Individual pixels are labeled by computing a weighted similarity metric of a pixel with all its neighbors, where the weights correspond to the neighboring pixel's strength. The neighbor that results in the largest weight greater than the given pixel's strength, confers its label to the given pixel. After the segmentation, the user can edit the segmentation by providing additional gestures in the image as illustrated in the figure below. |
Screen Captures
Click to enlarge photos if necessary
Similar Effects
See also the Fast GrowCut documentation. This is a fast version of the GrowCut algorithm. It is downloadable in the Extension Manager, thus requires memory.
References
- V. Vezhnevets and V. Konouchine, "GrowCut - Interactive multi-label N-D image segmentation", in Proc. Graphicon, 2005. pp. 150--156.