Difference between revisions of "Documentation/Nightly/Modules/PkModeling"
Stevedaxiao (talk | contribs) |
|||
| Line 68: | Line 68: | ||
* '''IO''' | * '''IO''' | ||
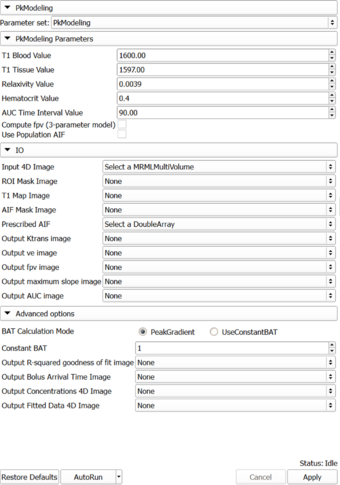
** '''Input 4D Image''': 4D DCE-MRI data | ** '''Input 4D Image''': 4D DCE-MRI data | ||
| + | ** '''T1 Map Image''': T1 Map | ||
** '''AIF Mask Image''': Mask designating the location of the arterial input function (AIF). AIF can either be calculated from the input using the aifMask, prescribed directly in concentration units using the prescribedAIF option, or via a population AIF. | ** '''AIF Mask Image''': Mask designating the location of the arterial input function (AIF). AIF can either be calculated from the input using the aifMask, prescribed directly in concentration units using the prescribedAIF option, or via a population AIF. | ||
** '''Prescribed AIF''': Prescribed arterial input function (AIF). AIF can either be calculated from the input using the aifMask option, via a population AIF, or can be prescribed directly in concentration units using the prescribedAIF option. An example of how a prescribed AIF can be defined is in [[File:AIF_example.mcsv.zip|this example .mcsv file]] (unzip before importing into Slicer!). Note that the x column corresponds to timestamps in seconds, and the y column is the contrast agent concentration (NOT image signal intensity). | ** '''Prescribed AIF''': Prescribed arterial input function (AIF). AIF can either be calculated from the input using the aifMask option, via a population AIF, or can be prescribed directly in concentration units using the prescribedAIF option. An example of how a prescribed AIF can be defined is in [[File:AIF_example.mcsv.zip|this example .mcsv file]] (unzip before importing into Slicer!). Note that the x column corresponds to timestamps in seconds, and the y column is the contrast agent concentration (NOT image signal intensity). | ||
| Line 76: | Line 77: | ||
** '''Output AUC image''': area under the curve in the first 90 seconds | ** '''Output AUC image''': area under the curve in the first 90 seconds | ||
* '''Advanced options''': | * '''Advanced options''': | ||
| + | ** '''BAT Calculation Mode''': PeakGradient (Default) or UseConstantBAT | ||
| + | ** '''Constant BAT''': Constant Bolus Arrival Time Index (frame number) | ||
** '''Output R-squared goodness of fit image''': each pixel will be initialized to a value between 0 and 1 characterizing the goodness of fit. Larger values correspond to a better fit (see [http://en.wikipedia.org/wiki/Coefficient_of_determination R^2 measure description]) | ** '''Output R-squared goodness of fit image''': each pixel will be initialized to a value between 0 and 1 characterizing the goodness of fit. Larger values correspond to a better fit (see [http://en.wikipedia.org/wiki/Coefficient_of_determination R^2 measure description]) | ||
** '''Output Bolus Arrival Time Image''': the bolus arrival time calculated at each pixel | ** '''Output Bolus Arrival Time Image''': the bolus arrival time calculated at each pixel | ||
Revision as of 21:26, 4 January 2016
Home < Documentation < Nightly < Modules < PkModeling
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: PkModeling | |||||||
|
Module Description
|
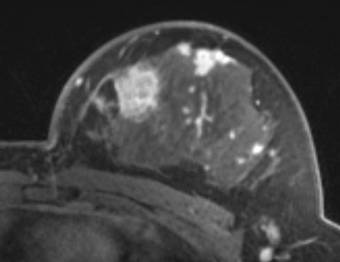
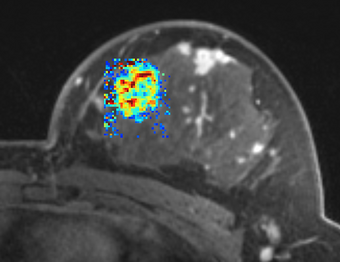
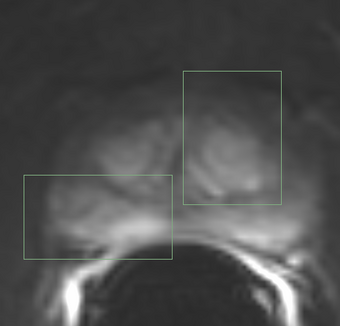
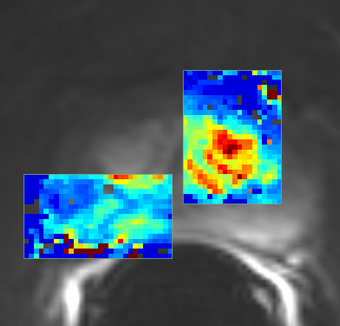
PkModeling (Pharmacokinetics Modeling) calculates quantitative parameters from Dynamic Contrast Enhanced DCE-MRI images. This module performs two operations:
|
Use Cases
- estimation of quantitative perfusion parameters from DCE MRI
- treatment response evaluation
- breast, prostate, brain DCE MRI analysis
Tutorials
Panels and their use
|
The following acquisition parameters should be available in the NRRD header of the input data (if you are analyzing a DICOM time series, they will typically be extracted from the DICOM data):
- TR Value: Repetition time (milliseconds)
- TE Value: Echo time (milliseconds)
- FA Value: Flip angle (degrees)
- Timestamps for the dynamic series (in milliseconds)
Here is an example how this information is represented in the NRRD header:
MultiVolume.DICOM.EchoTime:=2.93 MultiVolume.DICOM.FlipAngle:=10 MultiVolume.DICOM.RepetitionTime:=6.13 MultiVolume.FrameIdentifyingDICOMTagName:=AcquisitionTime MultiVolume.FrameIdentifyingDICOMTagUnits:=ms
Similar Modules
References
- [1] Knopp MV, Giesel FL, Marcos H et al: Dynamic contrast-enhanced magnetic resonance imaging in oncology. Top Magn Reson Imaging, 2001; 12:301-308.
- [2] Rijpkema M, Kaanders JHAM, Joosten FBM et al: Method for quantitative mapping of dynamic MRI contrast agent uptake in human tumors. J Magn Reson Imaging 2001; 14:457-463.
- [3] de Bazelaire, C.M., et al., MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3.0 T: preliminary results. Radiology, 2004. 230(3): p. 652-9.
- [4] Pintaske J, Martirosian P, Graf H, Erb G, Lodemann K-P, Claussen CD, Schick F. Relaxivity of Gadopentetate Dimeglumine (Magnevist), Gadobutrol (Gadovist), and Gadobenate Dimeglumine (MultiHance) in human blood plasma at 0.2, 1.5, and 3 Tesla. Investigative radiology. 2006 March;41(3):213–21.
- [5] Parker GJ, Roberts C, Macdonald A, Buonaccorsi GA, Cheung S, Buckley DL, Jackson A, Watson Y, Davies K, Jayson GC. Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magnetic Resonance in Medicine, 2006 Nov; 56(5):993-1000.
- [6] Huang, W., Li, X., Chen, Y., Li, X., Chang, M.-C., Oborski, M. J., … Kalpathy-Cramer, J. (2014). Variations of dynamic contrast-enhanced magnetic resonance imaging in evaluation of breast cancer therapy response: a multicenter data analysis challenge. Translational Oncology, 7(1), 153–66. doi:10.1593/tlo.13838 http://dx.doi.org/10.1593/tlo.13838
- [7] Tofts, P. S., Brix, G., Buckley, D. L., Evelhoch, J. L., Henderson, E., Knopp, M. V, … Weisskoff, R. M. (1999). Estimating kinetic parameters from Contrast-Enhanced T 1 -Weighted MRI of a Diffusable Tracer : Standardized Quantities and Symbols. J Magn Reson Imaging, 10(3), 223–232.
Information for Developers
| Section under construction. |
Source code: https://github.com/millerjv/PkModeling