Difference between revisions of "Documentation/4.4/gif tutorial v3 3"
From Slicer Wiki
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | ===Loading DTI and Baseline Data=== | + | |[[image:Neurosurgical Planning tutorial.png|700px|center]] |
| + | |||
| + | {| class="wikitable" style="border-spacing: 2px; border: 1px solid darkgray;" | ||
| + | ! scope="col" | Table of Contents | ||
| + | |- | ||
| + | |[[Documentation/4.4/gif tutorial v3 1|1. Loading DTI and Baseline Data]] | ||
| + | |- | ||
| + | |[[Documentation/4.4/gif tutorial v3 2|2. Segmentation]] | ||
| + | |- | ||
| + | |[[Documentation/4.4/gif tutorial v3 4|4. Tractography exploration of the ipsilateral and contralateral side]] | ||
| + | |} | ||
| + | |||
| + | ===Tractography exploration of peritumoral white matter fibers=== | ||
{| class="wikitable3" cellpadding="5px" | {| class="wikitable3" cellpadding="5px" | ||
| Line 6: | Line 18: | ||
|style="width:50%"|[[image:y.gif|frame|600px]] | |style="width:50%"|[[image:y.gif|frame|600px]] | ||
|-valign="top" | |-valign="top" | ||
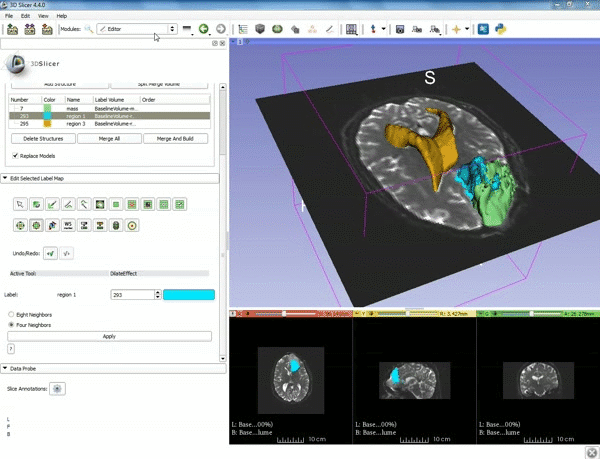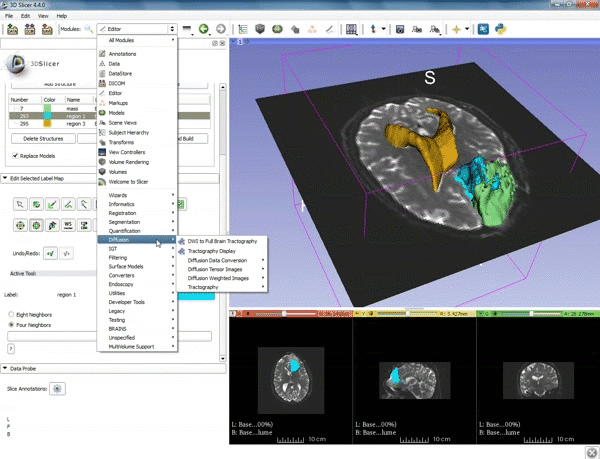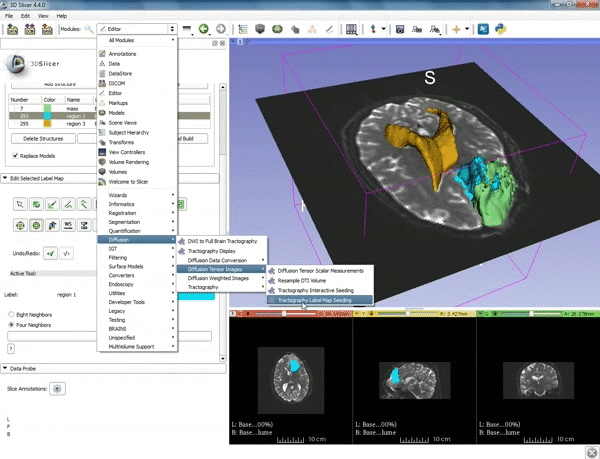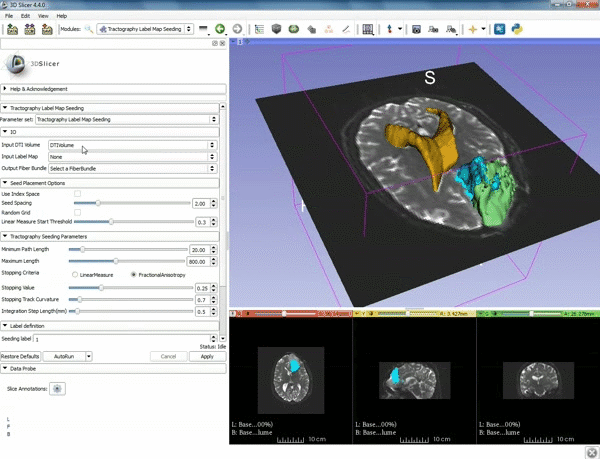
| − | |1. a) Make sure the cystic part is selected and visible in the 3 slice views. b) Click on the Dilate Effect tool then click on the cystic part of the tumor on the red slice view. | + | |1. a) Make sure the cystic part is selected and visible in the 3 slice views. b) Click on the Dilate Effect tool then click on the cystic part of the tumor on the red slice view. |
|2. Click on Apply 3 times to generate the peritumoral volume. | |2. Click on Apply 3 times to generate the peritumoral volume. | ||
|-align="center" | |-align="center" | ||
| Line 18: | Line 30: | ||
|style="width:50%"|[[image:b.gif|frame|600px]] | |style="width:50%"|[[image:b.gif|frame|600px]] | ||
|-valign="top" | |-valign="top" | ||
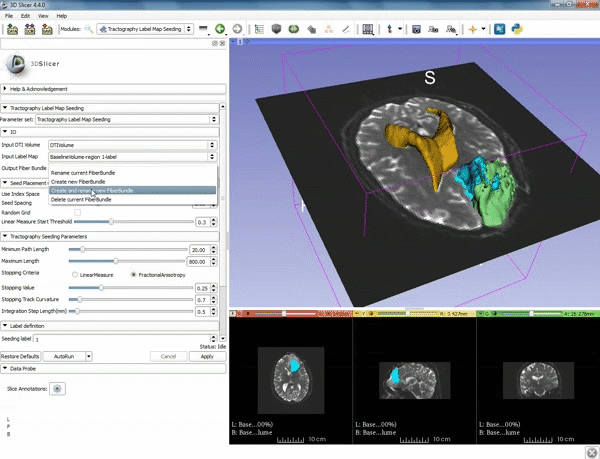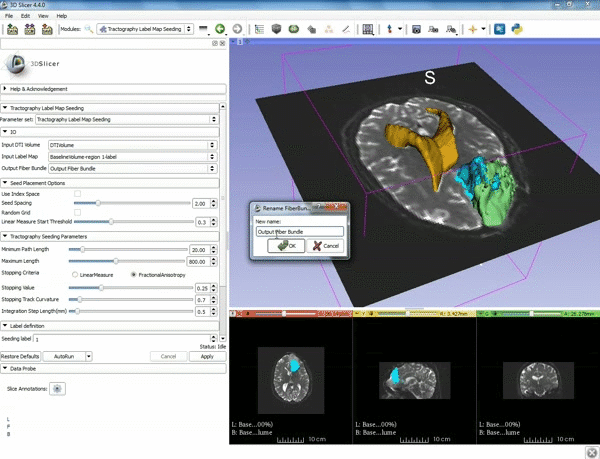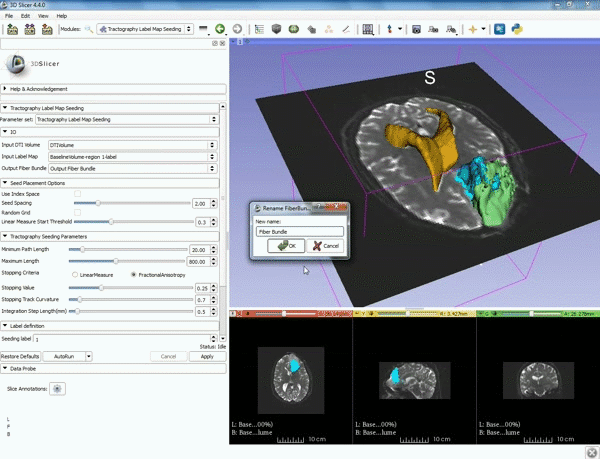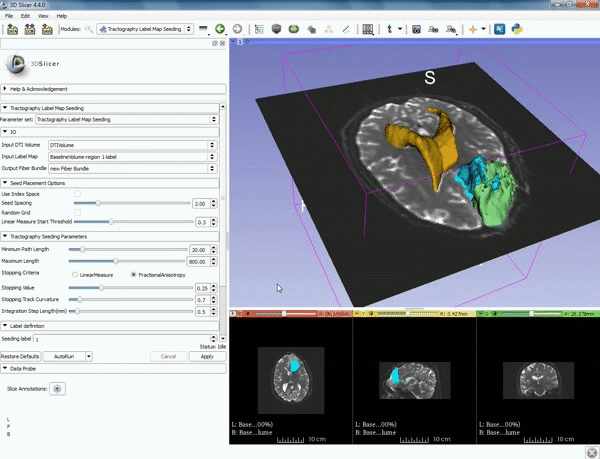
| − | |5. a) For Output Fiber Bundle select "Create and rename newFiberBundle". b)When the pop-up appears rename the fiber bundle to to "newFiberBundle". | + | |5. a) For Output Fiber Bundle select "Create and rename newFiberBundle". b)When the pop-up appears rename the fiber bundle to to "newFiberBundle". |
| − | |6.a) Under the Seed Placement Options: Check the Use Index Space. b) Under Tractography Seeding parameter set the stopping value to "0.15" and make sure for Stopping Criteria that "Fractional Anisotropy" is selected. | + | |6. a) Under the Seed Placement Options: Check the Use Index Space. b) Under Tractography Seeding parameter set the stopping value to "0.15" and make sure for Stopping Criteria that "Fractional Anisotropy" is selected. |
|-align="center" | |-align="center" | ||
|style="width:50%"|[[image:d.gif|frame|600px]] | |style="width:50%"|[[image:d.gif|frame|600px]] | ||
| Line 25: | Line 37: | ||
|7. Under Label Definition set the "Seeding Label" to 293, and Click on Apply. | |7. Under Label Definition set the "Seeding Label" to 293, and Click on Apply. | ||
|} | |} | ||
| + | |||
| + | |||
| + | |||
| + | <div style="text-align: center;">'''[[Documentation/4.4/gif tutorial v3 4|Next Step]]'''</div> | ||
| + | <div style="text-align: center;">'''[[Documentation/4.4/gif tutorial v3 2|Previous Step]]'''</div> | ||
Latest revision as of 20:28, 20 July 2015
Home < Documentation < 4.4 < gif tutorial v3 3|
| Table of Contents |
|---|
| 1. Loading DTI and Baseline Data |
| 2. Segmentation |
| 4. Tractography exploration of the ipsilateral and contralateral side |
Tractography exploration of peritumoral white matter fibers
| 1. a) Make sure the cystic part is selected and visible in the 3 slice views. b) Click on the Dilate Effect tool then click on the cystic part of the tumor on the red slice view. | 2. Click on Apply 3 times to generate the peritumoral volume. |
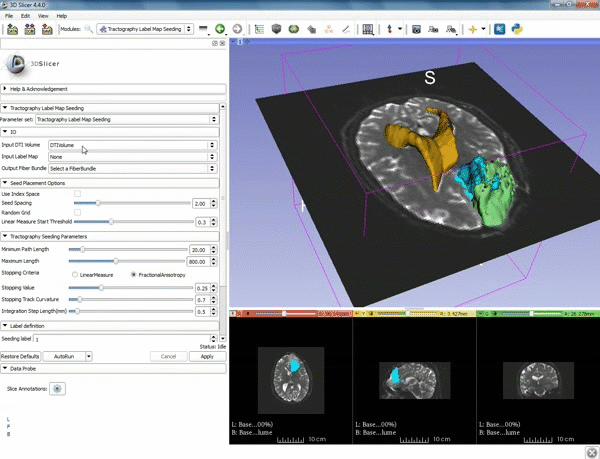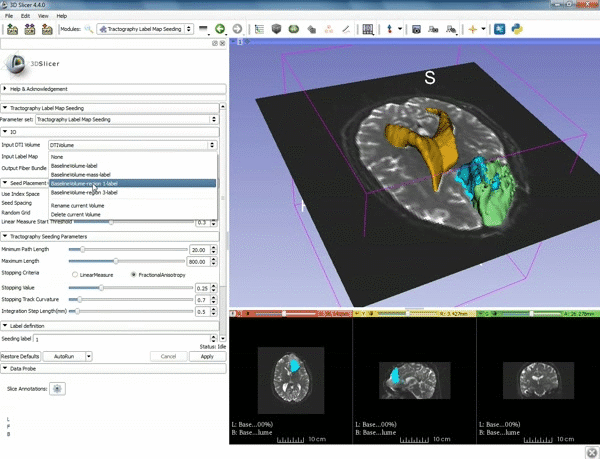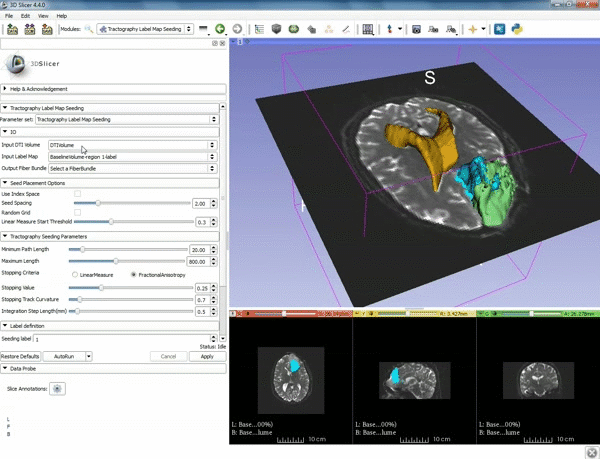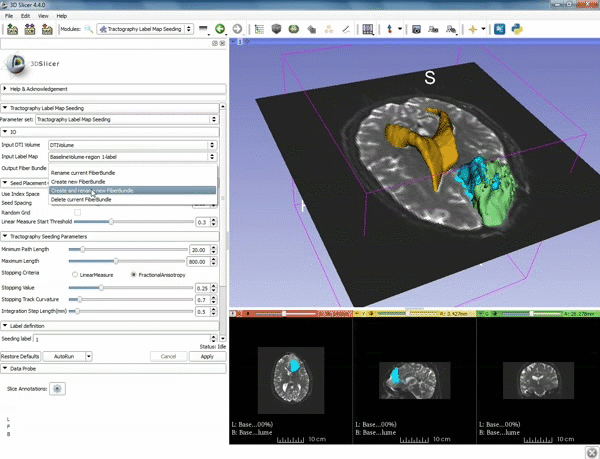
| 3. Select the module "Tractography Label Map Seeding" by clicking "Editor" at the top, then "Diffusion", then "Diffusion Tensor Imaging". | 4. Make sure that "DTI Volume" is selected for Input DTI Volume and "BaselineVolumeregion_1-label" is selected for Input Label Map. |
| 5. a) For Output Fiber Bundle select "Create and rename newFiberBundle". b)When the pop-up appears rename the fiber bundle to to "newFiberBundle". | 6. a) Under the Seed Placement Options: Check the Use Index Space. b) Under Tractography Seeding parameter set the stopping value to "0.15" and make sure for Stopping Criteria that "Fractional Anisotropy" is selected. |
| 7. Under Label Definition set the "Seeding Label" to 293, and Click on Apply. |