Difference between revisions of "Documentation/Nightly/Modules/T1Mapping"
Stevedaxiao (talk | contribs) (Created page with '<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- -----------------------…') |
Stevedaxiao (talk | contribs) |
||
| Line 49: | Line 49: | ||
| | | | ||
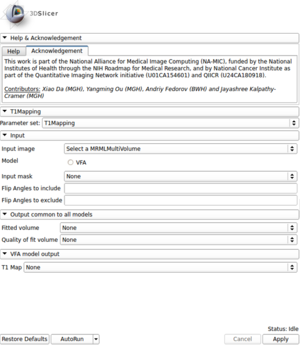
*'''Input''' | *'''Input''' | ||
| − | ** '''Input image''': | + | ** '''Input image''': Multivolume node containing multiple flip angles image loaded using DICOM module |
** '''Model''': VFA, Variable Flip Angle | ** '''Model''': VFA, Variable Flip Angle | ||
| − | ** '''Input mask''': | + | ** '''Input mask''': Segmentation of the region of interest (optional); if specified, model fitting will be performed only within the specified mask |
| − | ** '''Flip Angles to include''': | + | ** '''Flip Angles to include''': List of Flip Angles that should be used in the fitting process (optional); this parameter is used only if not empty |
| − | ** '''Flip Angles to exclude''': | + | ** '''Flip Angles to exclude''': List of Flip Angles that should NOT be used in the fitting process (optional); this parameter is used only if not empty |
*'''Output common to all models''' | *'''Output common to all models''' | ||
| − | ** '''Fitted volume''': | + | ** '''Fitted volume''': Multi-volume containing the fitted model sampled at the Flip Angles of the input dataset (optional) |
| − | ** '''Quality of fit volume''': | + | ** '''Quality of fit volume''': R-squared (optional |
*'''VFA model output''' | *'''VFA model output''' | ||
Revision as of 18:05, 15 July 2015
Home < Documentation < Nightly < Modules < T1Mapping
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: T1_Mapping_CPP | |||||||
|
Module Description
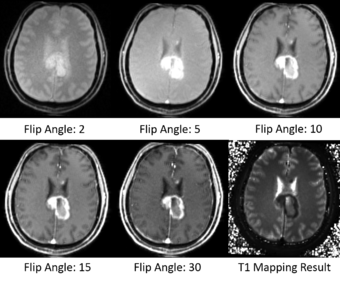
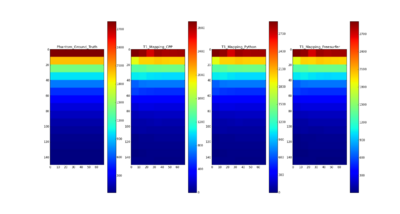
T1 mapping estimates effective tissue parameter maps (T1) from multi-spectral FLASH MRI scans with different flip angles. T1 mapping can be used to optimize parameters for a sequence, monitor diseased tissue, measure Ktrans in DCE-MRI and etc.
The method and equations for Variable Flip Angle T1 Mapping are available here: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3620726/pdf/nihms-423474.pdf
Use Cases
- Take multi-spectral FLASH images with an arbitrary number of flip angles as input, and estimate the T1 values of the data for each voxel.
- Read repetition time(TR), echo time(TE) and flip angles from the Dicom header directly.
- Prostate, brain, head & neck, cervix, breast and etc.
Tutorials
Panels and their use
|
Similar Modules
References
Information for Developers
| Section under construction. |
Source code: https://github.com/stevedaxiao/T1_Mapping_CPP