Difference between revisions of "Documentation/Nightly/Extensions/TCIABrowser"
| Line 75: | Line 75: | ||
By opening TCIABrowser module it will automatically connect to the the TCIA server and list all of the available collections. | By opening TCIABrowser module it will automatically connect to the the TCIA server and list all of the available collections. | ||
| − | |||
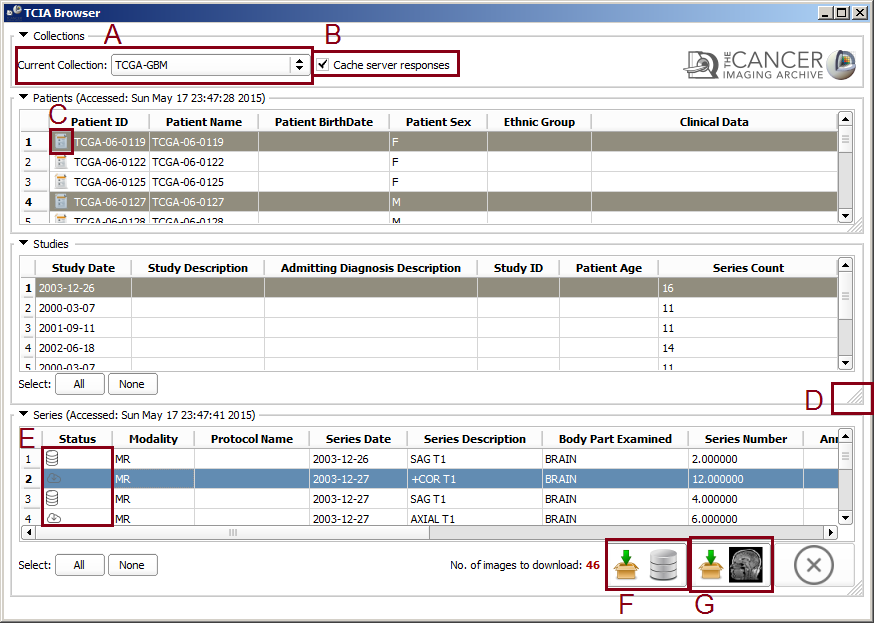
In the browser window from the combo box select a collection. The browser will get the patient data from the TCIA server and will populate the Patients table. In patient table, the clinical data icon besides the patient ID [[Image:Report-icon-tcia.png|thumb|50px|Clinical Data Icon]] means that the clinical data for that specific patient is available in TCGA. The use Cache check box will cache the query results on your hard drive which makes the next selections much faster. If you want to do the query (connect to TCIA Server) you can uncheck this box. Above the Patients table you can see the last access time. The process is the same for studies and series tables, selecting a patient will populate the study table for the selected one and selecting a study will update the series table. | In the browser window from the combo box select a collection. The browser will get the patient data from the TCIA server and will populate the Patients table. In patient table, the clinical data icon besides the patient ID [[Image:Report-icon-tcia.png|thumb|50px|Clinical Data Icon]] means that the clinical data for that specific patient is available in TCGA. The use Cache check box will cache the query results on your hard drive which makes the next selections much faster. If you want to do the query (connect to TCIA Server) you can uncheck this box. Above the Patients table you can see the last access time. The process is the same for studies and series tables, selecting a patient will populate the study table for the selected one and selecting a study will update the series table. | ||
| − | |[[ | + | |[[File:TCIA-Browser-1.png|thumb|6000px|Module GUI]] |
| + | |||
| + | |||
|} | |} | ||
Revision as of 13:22, 18 May 2015
Home < Documentation < Nightly < Extensions < TCIABrowser
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: TCIABrowser License: Slicer License
|
Module Description
The Cancer Imaging Archive (TCIA) hosts a large collection of Cancer medical imaging data which is available to public through a programmatic interface (REST API). TCIA Browser is a Slicer module by which the user can connect to the TCIA archive, browse different collections, patient subjects, studies and series, download the images and visualize them in 3D Slicer.
As of February 2014, the following collections of TCIA are available via public TCIA API interface (see description of TCIA collections for details):
- QIBA CT-1C
- TCGA-GBM *
- RIDER PHANTOM PET-CT
- RIDER PHANTOM MRI
- Phantom FDA
- TCGA-LGG *
- TCGA-KIRC *
- TCGA-BRCA *
- BREAST-DIAGNOSIS
- PROSTATE-DIAGNOSIS
- LIDC-IDRI
- TCGA-LUAD *
- RIDER Lung CT
- RIDER NEURO MRI
- RIDER Lung PET-CT
- CT COLONOGRAPHY
- REMBRANDT
- RIDER Breast MRI
For the collections marked with *, available clinical data is linked from The Cancer Genome Atlas.
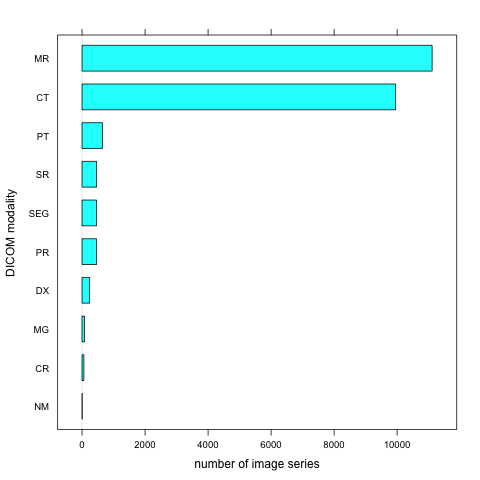
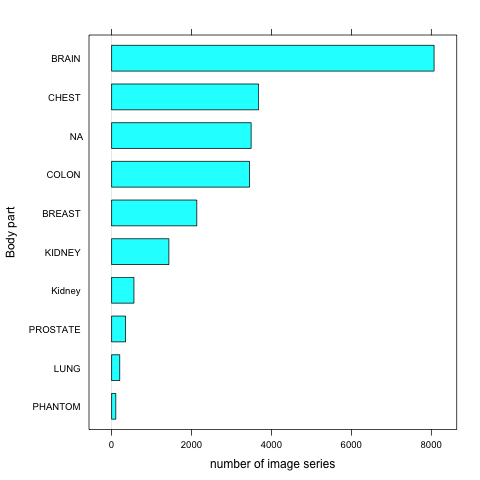
TCIABrowser gives you access to 18 collections, which contain a total of over 23,000 image series from a variety of manufacturers and of different modalities and organs. Some of the summary statistics of the data available from the public collections listed is provided below.
Tutorials
Not available at this time.
Panels and their use
- Browsing Collections, patients and studies
|
|
- Downloading Series
- Access Clinical Data from TCGA
- Settings
Similar Modules
References
- Quantitative Image Informatics for Cancer Research (QIICR)
- Quantitative Imaging Network (QIN)
- TCIA Home Page
- cBioPortal for Cancer Genomics Web Interface
- Description of TCIA Collections
- TCIA Rest API Documentation
- Project page at NAMIC 2014 Project Week
- Mashape page for testing TCIA API endpoint
Information for Developers
Tip: The REST API can be checked by REST Console Google Chrome extension.
| Section under construction. |