Difference between revisions of "Documentation/4.3/Modules/DoseComparison"
(Nightly -> 4.3) |
|||
| Line 30: | Line 30: | ||
{|align="center" | {|align="center" | ||
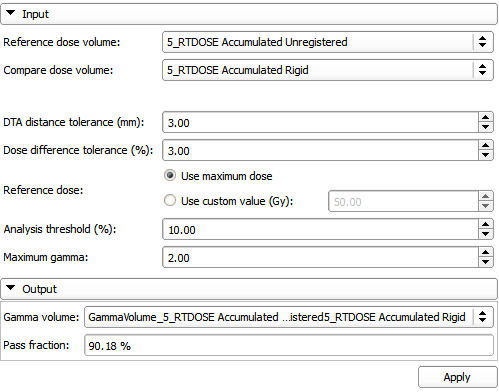
| − | |[[File: | + | |[[File:SlicerRT_0.12_DoseComparisonModuleUi.png|thumb|499px|Dose Comparison module UI]] |
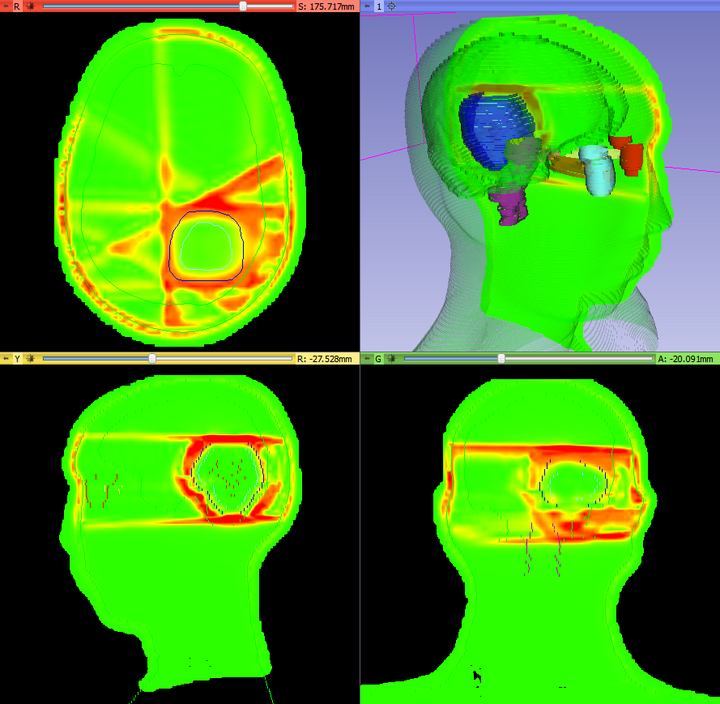
| − | |[[File: | + | |[[File:SlicerRT_0.11_Gamma.png|thumb|720px|Example of the result gamma volume]] |
|} | |} | ||
Revision as of 17:49, 5 November 2013
Home < Documentation < 4.3 < Modules < DoseComparison
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is part of the SparKit project, funded by An Applied Cancer Research Unit of Cancer Care Ontario with funds provided by the Ministry of Health and Long-Term Care and the Ontario Consortium for Adaptive Interventions in Radiation Oncology (OCAIRO) to provide free, open-source toolset for radiotherapy and related image-guided interventions. | |||||||
|
Module Description
The DoseComparison module computes the difference between two co-registered dose volumes using the gamma dose distribution comparison method. This module uses the gamma Plastimatch algorithm internally.
Use Cases
Comparing dose distributions
Tutorials
Panels and their use
- Current contour(s): Currently selected contour node (vtkMRMLContourNode) or conotur hierarchy node (vtkMRMLContourHierarchyNode)
- Active representation: The active representation (Ribbon model / Indexed labelmap / Closed surface model) of the selected contour. If a hierarchy is selected in which the active representations of the contained contours do not match, Various is indicated
- Change active representation: This box contains controls to convert from one representation to another
Similar Modules
References
- Low, D. A.; Harms, W. B.; Mutic, S. & Purdy, J. A. A technique for the quantitative evaluation of dose distributions Med. Phys, 1998, 25 (5), 656/6
- Low, D. A. & Dempsey, J. F. Evaluation of the gamma dose distribution comparison method Med. Phys, 2003, 30 (9), 2455/10
Information for Developers
N/A