Difference between revisions of "Slicer4:MultiVolumeContainer"
From Slicer Wiki
| (10 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[Slicer4:Developers#Projects|Back to Slicer4 Projects]] | [[Slicer4:Developers#Projects|Back to Slicer4 Projects]] | ||
| + | |||
| + | <gallery widths=400px heights=300px> | ||
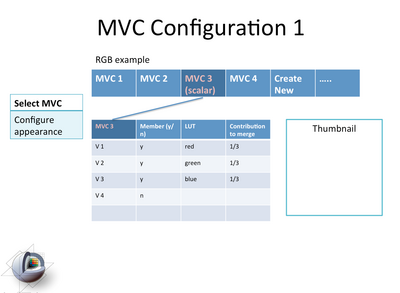
| + | image:Slicer4-MVC1Module-Mockup-2011-06-27.png|Mockup of the functionality in the proposed MVC module. [[media:ViewingModules-2011-06-27.ppt|See here for the ppt]] | ||
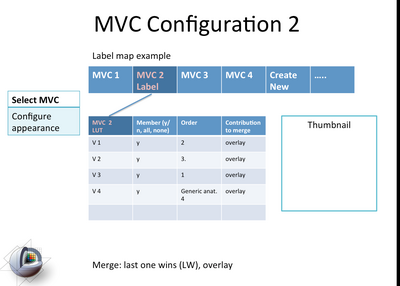
| + | image:Slicer4-MVC2Module-Mockup-2011-06-27.png|Mockup of the functionality in the proposed MVC module. [[media:ViewingModules-2011-06-27.ppt|See here for the ppt]] | ||
| + | </gallery> | ||
=Introduction= | =Introduction= | ||
| Line 62: | Line 67: | ||
==Meeting Notes== | ==Meeting Notes== | ||
| + | |||
| + | * Use cases | ||
| + | ** DWI | ||
| + | ** longitudinal assessments | ||
| + | ** DCE | ||
| + | ** fMRI | ||
| + | ** atlas visualisation/exploration (sets of images from different patients) | ||
| + | *4D Image module in Slicer4 | ||
| + | ** too many volume nodes in the node selectors | ||
| + | ** copy of the image data into a "front" reference volume for volume rendering | ||
| + | * want multiple dimensions/parameters | ||
| + | * Proposed implementations: | ||
| + | ** one volume with multiple vtkImageData | ||
| + | *** use deep copy, keep the ijk to ras matrix, doesn't have to be same size | ||
| + | *** set and observe? | ||
| + | ** volume hierarchies, closed by default? | ||
| + | ** switch node | ||
| + | ** load on demand | ||
| + | ** back end database | ||
| + | * concept of derived data, volumes associated with a base volume | ||
| + | * creating a new multi volume from loaded volumes: designate it as a time series or some other logical grouping and tools could target them | ||
| + | * save a mutivolume container as an entity | ||
| + | ** nrrd can't handle it, nor nifti, as one file | ||
| + | ** mrml could encapsulate it - multiple storage nodes | ||
| + | * be able to view/compare 1,2,3 volumes at a time | ||
| + | * for hierarchies, implement as much as possible on the superclass (ie only show this volume if part of a group, iterate over all volumes, etc) | ||
| + | * user hierarchies to group, but also have a multi volume node that can swap out vtkImageData | ||
| + | * for IGT want to be able to inspect last 10-15 frames with label maps, moving set of recent volumes | ||
| + | * vtkImageStash used for undo on Editor | ||
| + | * granularity of time points, let the user determine what's one time point without ref do timestamp | ||
| + | * if just used hierarchies | ||
| + | ** user interface issues to group them | ||
| + | ** load on demand so don't overwhelm memory | ||
| + | ** display node on the top level hierarchy node | ||
| + | * volumes with a vector defined at each voxel, so each volume has the same size | ||
| + | ** DWI is about angle not time as the parameter over which to iterate | ||
| + | * can have a time series of multichannel data | ||
| + | * run time generality versus class definitions | ||
| + | * add a frame number to say that this can have multiple internal elements | ||
| + | * Ron's motiviation: be able to say what changed from previous time point | ||
| + | ** is an argument to not deal with fMRI, DWI | ||
| + | ** by the end of 2011 have a prototype so can push this in grant writing | ||
| + | * Andrey has a stack of scans at time points, different positions, across an intervention, currently can't accomodate the sorting by time points | ||
| + | * need to be able to put one volume in multiple groups | ||
| + | * time stamp at vtkMRMLNodeLevel? | ||
| + | ** mrml displayable hierarchy logic to provide helper classes (reorder children by time) | ||
| + | * prototype it in the dicom query/retrieve IGT project | ||
| + | ** just hierarchies | ||
| + | ** up to 50 time points, up to 50 volumes per time point | ||
| + | ** demo at Jan 2012 project | ||
| + | |||
| + | === Follow up discussion === | ||
| + | |||
| + | Nicole, Wendy and Steve talked about mrml structure in Slicer. | ||
| + | |||
| + | * When read in a DICOM file, create a hierarchy like: | ||
| + | **patient | ||
| + | ***study0 | ||
| + | ****series0 is equivalent to a single volume0 | ||
| + | ****series1 is equivalent to a single volume1 | ||
| + | ***study1 | ||
| + | ****series0 is equivalent to a single multivolume0 | ||
| + | * When create some derived data can end up with a mrml scene that looks like: | ||
| + | **patient | ||
| + | ***study0 | ||
| + | ****series0 is equivalent to a single volume0 | ||
| + | ****series1 is equivalent to a single volume1 | ||
| + | *****labelmap1 is derived from volume1 | ||
| + | ******model1 is derived from labelmap1 | ||
| + | ***study1 | ||
| + | ****series0 is equivalent to a single multivolume0 | ||
| + | *****a parameter map dervied from the multivolume0 | ||
| + | * first get the dicom to mrml hierarchy creation automatic, then provide an interface to build one via drag and drop, then make sure that derived data is associated with the correct nodes | ||
| + | ** see the top of the page for a mock-up of the interface. | ||
Latest revision as of 14:16, 27 June 2011
Home < Slicer4:MultiVolumeContainerBack to Slicer4 Projects
Mockup of the functionality in the proposed MVC module. See here for the ppt
Mockup of the functionality in the proposed MVC module. See here for the ppt
Contents
Introduction
This page describes the concept for an API for time series management. What is needed is an image informatics frame work to organize, manage, and analyze multi volume data inside Slicer. If successful, this will allow more user-friendlyness.
Examples
- R: RECIST analysis
- M: Time Series Analysis of MRI's from an MS Patient
- P: Time Series Analysis of SUV values in MR/CT PET
- this has wider applicability for calculation of the changes in statistics of a certain parameter over an ROI (example: pharmacokinetic parameter, T2 value or ADC for monitoring treatment in prostate cancer)
- D: Dynamic Contrast Enhancement (DCE)
- T: NA-MIC TBI: multichannel multi time
Common Features and Needs
- These are all time series of scalar volumes and need to be organized in a number of dimensions: space, time, signal intensities, feature space (such as channels in multichannel data, gradient strength in dMRI data, etc.)
- Typically the volumes need to be registered to each other in order to organize them in space: Fusion between the channels, fusion across the time points
- bias correction is often needed: MR, PET, CT(?)
- Window/Level:
- Allow manual setting of w/l for the reference time point.
- CT: one w/l for all time points, PET, MR: the SUV concept could be expanded to MRI: use one or two user selected/drawn ROI to adjust window level across time points.
Viewing and Analysis
- view across time: movie (loop, bounce)
- graphing: resample across time
- need to do analysis across time: ROI's (label map) for voxel statistics across time. Create spreadsheet / graph as result.
- need to store the organizational frame work. When the patient comes for the next scan we need ability to add a time point, fix data (registration, w/l, update spreadsheet/graph)
- need to calculate statistics over time for several volumes at each time point (motivating application: multiparametric MRI for treatment monitoring, PI F.Fennessy)
Workflows
Initial Setup of a MultiVolumeContainer
- Assume that all data have been loaded into Slicer already
- Create a new MultiVolumeContainer.
- MVC's are always multi-channel. Single channel is just a special case.
- User specifies the number of channels.
- one of the volumes is assigned as the reference volume (default is the first loaded, user can change manually by clicking a checkbox)
Things you can do with MVC's
- editing the MVC by adding or removing time points or changing their order
- change the reference volume
- assign registration types between the channels and between the time points
- execute the registration cascade
- adjust W/L across time for each channel using histogram equalization with manual override
- across time analysis: time versus voxel-statistics or dimensions, static, interactive adjustments
- special case for SUV across time
- RECIST: determination at each time point, use previous one as initialization
- Modeling: In addition to time versus voxel-statistics type of curves sometimes there is a need for modeling and parameter extraction.
- DCE requires computation of the Tofts model: a curve is fitted to each voxel time course and different parameters are extracted per voxel.
- NukMed uses other, similar formulas,
- DWI to DTI uses the Stejskal formula (or similar). The resulting tensor fields are then used for further post-processing.
Implementation in Slicer 4
Steve Pieper: It seems that the generalized hiearchies in Slicer4 will be the right underlying layer
Summer 2011 Project Week Breakout Session
Attendees:
Agenda:
- Define use cases
- List possible implementation design
- Decide on an implementation design
Meeting Notes
- Use cases
- DWI
- longitudinal assessments
- DCE
- fMRI
- atlas visualisation/exploration (sets of images from different patients)
- 4D Image module in Slicer4
- too many volume nodes in the node selectors
- copy of the image data into a "front" reference volume for volume rendering
- want multiple dimensions/parameters
- Proposed implementations:
- one volume with multiple vtkImageData
- use deep copy, keep the ijk to ras matrix, doesn't have to be same size
- set and observe?
- volume hierarchies, closed by default?
- switch node
- load on demand
- back end database
- one volume with multiple vtkImageData
- concept of derived data, volumes associated with a base volume
- creating a new multi volume from loaded volumes: designate it as a time series or some other logical grouping and tools could target them
- save a mutivolume container as an entity
- nrrd can't handle it, nor nifti, as one file
- mrml could encapsulate it - multiple storage nodes
- be able to view/compare 1,2,3 volumes at a time
- for hierarchies, implement as much as possible on the superclass (ie only show this volume if part of a group, iterate over all volumes, etc)
- user hierarchies to group, but also have a multi volume node that can swap out vtkImageData
- for IGT want to be able to inspect last 10-15 frames with label maps, moving set of recent volumes
- vtkImageStash used for undo on Editor
- granularity of time points, let the user determine what's one time point without ref do timestamp
- if just used hierarchies
- user interface issues to group them
- load on demand so don't overwhelm memory
- display node on the top level hierarchy node
- volumes with a vector defined at each voxel, so each volume has the same size
- DWI is about angle not time as the parameter over which to iterate
- can have a time series of multichannel data
- run time generality versus class definitions
- add a frame number to say that this can have multiple internal elements
- Ron's motiviation: be able to say what changed from previous time point
- is an argument to not deal with fMRI, DWI
- by the end of 2011 have a prototype so can push this in grant writing
- Andrey has a stack of scans at time points, different positions, across an intervention, currently can't accomodate the sorting by time points
- need to be able to put one volume in multiple groups
- time stamp at vtkMRMLNodeLevel?
- mrml displayable hierarchy logic to provide helper classes (reorder children by time)
- prototype it in the dicom query/retrieve IGT project
- just hierarchies
- up to 50 time points, up to 50 volumes per time point
- demo at Jan 2012 project
Follow up discussion
Nicole, Wendy and Steve talked about mrml structure in Slicer.
- When read in a DICOM file, create a hierarchy like:
- patient
- study0
- series0 is equivalent to a single volume0
- series1 is equivalent to a single volume1
- study1
- series0 is equivalent to a single multivolume0
- study0
- patient
- When create some derived data can end up with a mrml scene that looks like:
- patient
- study0
- series0 is equivalent to a single volume0
- series1 is equivalent to a single volume1
- labelmap1 is derived from volume1
- model1 is derived from labelmap1
- labelmap1 is derived from volume1
- study1
- series0 is equivalent to a single multivolume0
- a parameter map dervied from the multivolume0
- series0 is equivalent to a single multivolume0
- study0
- patient
- first get the dicom to mrml hierarchy creation automatic, then provide an interface to build one via drag and drop, then make sure that derived data is associated with the correct nodes
- see the top of the page for a mock-up of the interface.