Difference between revisions of "EMSegmenter-Tasks:Non-Human-Primate"
From Slicer Wiki
Belhachemi (talk | contribs) |
Belhachemi (talk | contribs) |
||
| Line 1: | Line 1: | ||
| − | + | =Description= | |
Non Human Primate pipeline ...'''TODO''' | Non Human Primate pipeline ...'''TODO''' | ||
| − | + | =Anatomical Tree= | |
* Root | * Root | ||
| Line 24: | Line 24: | ||
*** background (BG) | *** background (BG) | ||
| − | + | =Atlas= | |
Image Dimension = 256 x 256 x 252 <br> | Image Dimension = 256 x 256 x 252 <br> | ||
Image Spacing = 0.4688 x 0.4688 x 0.50002 | Image Spacing = 0.4688 x 0.4688 x 0.50002 | ||
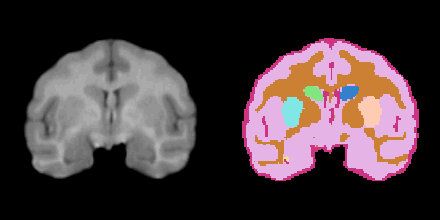
| − | + | =Result= | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
[[Image:NHP.png]] | [[Image:NHP.png]] | ||
| − | + | =Collaborators= | |
Andriy Fedorov (BWH) | Andriy Fedorov (BWH) | ||
| + | |||
| + | =Citations= | ||
| + | * Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC N4ITK: Improved N3 Bias Correction, IEEE Trans Med Imag, 2010 | ||
| + | * Pohl K, Bouix S, Nakamura M, Rohlfing T, McCarley R, Kikinis R, Grimson W, Shenton M, Wells W. [http://www.slicer.org/pages/Special:PubDB_View?dspaceid=608 A Hierarchical Algorithm for MR Brain Image Parcellation.] IEEE Transactions on Medical Imaging. 2007 Sept;26(9):1201-1212. | ||
| + | * S. Warfield, J. Rexilius, P. Huppi, T. Inder, E. Miller, W. Wells, G. Zientara, F. Jolesz, and R. Kikinis, “A binary entropy measure to assess nonrigid registration algorithms,” in MICCAI, LNCS, pp. 266–274, Springer, October 2001. | ||
| + | * Johnson H.J., Harris G., Williams K. [http://hdl.handle.net/1926/1291 BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit], The Insight Journal, July 2007 | ||
Revision as of 21:53, 1 December 2010
Home < EMSegmenter-Tasks:Non-Human-PrimateDescription
Non Human Primate pipeline ...TODO
Anatomical Tree
- Root
- Head
- intracranial cavity (ICC)
- TODO (WGC)
- grey matter (GM)
- TODO (lSGM)
- left caudate (lCaudate)
- left putamen (lPutamen)
- left hippocampus (lHippocampus)
- TODO (rSGM)
- right caudate (rCaudate)
- right putamen (rPutamen)
- right hippocampus (rHippocampus)
- TODO (CGM)
- TODO (lSGM)
- white matter (WM)
- cerebrospinal fluid (CSF)
- grey matter (GM)
- Brainstem
- Cerebellum
- TODO (WGC)
- background (BG)
- intracranial cavity (ICC)
- Head
Atlas
Image Dimension = 256 x 256 x 252
Image Spacing = 0.4688 x 0.4688 x 0.50002
Result
Collaborators
Andriy Fedorov (BWH)
Citations
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC N4ITK: Improved N3 Bias Correction, IEEE Trans Med Imag, 2010
- Pohl K, Bouix S, Nakamura M, Rohlfing T, McCarley R, Kikinis R, Grimson W, Shenton M, Wells W. A Hierarchical Algorithm for MR Brain Image Parcellation. IEEE Transactions on Medical Imaging. 2007 Sept;26(9):1201-1212.
- S. Warfield, J. Rexilius, P. Huppi, T. Inder, E. Miller, W. Wells, G. Zientara, F. Jolesz, and R. Kikinis, “A binary entropy measure to assess nonrigid registration algorithms,” in MICCAI, LNCS, pp. 266–274, Springer, October 2001.
- Johnson H.J., Harris G., Williams K. BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit, The Insight Journal, July 2007