Difference between revisions of "EMSegmenter-Tasks:MRI-Human-Brain"
From Slicer Wiki
| Line 3: | Line 3: | ||
==Description== | ==Description== | ||
Single channel automatic segmentation of t1w-MRI brain scans into the major tissue classes (gray matter, white matter, csf). The task can only be applied to t1w brain scan showing parts of the skull and neck. The pipeline consist of the following steps: | Single channel automatic segmentation of t1w-MRI brain scans into the major tissue classes (gray matter, white matter, csf). The task can only be applied to t1w brain scan showing parts of the skull and neck. The pipeline consist of the following steps: | ||
| − | * Step 1: Perform image inhomogeneity correction of the MRI scan via [http://www.slicer.org/slicerWiki/index.php/Modules:N4ITKBiasFieldCorrection-Documentation-3.6 N4ITKBiasFieldCorrection] (et al | + | * Step 1: Perform image inhomogeneity correction of the MRI scan via [http://www.slicer.org/slicerWiki/index.php/Modules:N4ITKBiasFieldCorrection-Documentation-3.6 N4ITKBiasFieldCorrection] (Tustison et al 2010) |
| − | * Step 2: Register the atlas to the MRI scan via [http://www.slicer.org/slicerWiki/index.php/Modules:BRAINSFit BRAINSFit] ( | + | * Step 2: Register the atlas to the MRI scan via [http://www.slicer.org/slicerWiki/index.php/Modules:BRAINSFit BRAINSFit] (ask Hans for citation) |
* Step 3: Compute the intensity distributions for each structure | * Step 3: Compute the intensity distributions for each structure | ||
| − | * Step 4: Automatically segment the MRI scan into the structures of interest using the EM Algorithm (et al | + | * Step 4: Automatically segment the MRI scan into the structures of interest using the EM Algorithm (Pohl et al 2007) |
==Anatomical Tree== | ==Anatomical Tree== | ||
Revision as of 18:00, 30 November 2010
Home < EMSegmenter-Tasks:MRI-Human-BrainMRI Human Brain
Description
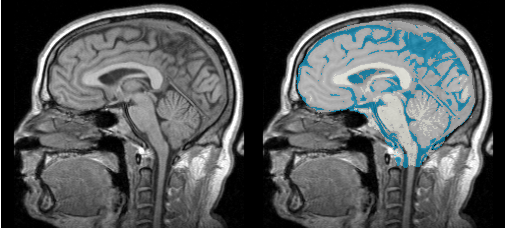
Single channel automatic segmentation of t1w-MRI brain scans into the major tissue classes (gray matter, white matter, csf). The task can only be applied to t1w brain scan showing parts of the skull and neck. The pipeline consist of the following steps:
- Step 1: Perform image inhomogeneity correction of the MRI scan via N4ITKBiasFieldCorrection (Tustison et al 2010)
- Step 2: Register the atlas to the MRI scan via BRAINSFit (ask Hans for citation)
- Step 3: Compute the intensity distributions for each structure
- Step 4: Automatically segment the MRI scan into the structures of interest using the EM Algorithm (Pohl et al 2007)
Anatomical Tree
- root
- background (BG)
- air (AIR)
- skull (skull)
- intracranial cavity (ICC)
- white matter (WM)
- grey matter (GM)
- cerebrospinal fluid (CSF)
- background (BG)
Atlas
Image Dimension = 256 x 256 x 124
Image Spacing = 0.9375 x 0.9375 x 0.1.5
Pre-Processing
Result
Citations
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC N4ITK: Improved N3 Bias Correction, IEEE Trans Med Imag, 2010
[3]