Difference between revisions of "Modules:GyriContourSegmentation-Documentation-3.5"
| (19 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | [[Documentation-3. | + | [[Documentation-3.6|Return to Slicer 3.5 Documentation]] |
__NOTOC__ | __NOTOC__ | ||
| − | === | + | ===Mesh Contour Segmentation=== |
MyModule | MyModule | ||
{| | {| | ||
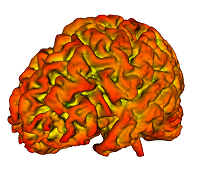
| − | |[[Image: | + | |[[Image:brainH.png|thumb|280px|Surface Normal Curvature]] |
| − | |[[Image: | + | |[[Image:tumor2_init_4.PNG|thumb|280px|Initial Points From User]] |
| − | |[[Image: | + | |[[Image:tumor_conv_3.png|thumb|280px|Converged Contour on Surface]] |
|} | |} | ||
| Line 25: | Line 25: | ||
===Module Description=== | ===Module Description=== | ||
| − | This module | + | This module is a tool to generate closed contours on a surface in 3D. The contour is initialized with a set of points, and subsequently 'evolves' according to some geometric criterion of the underlying surface (e.g. Surface Normal, mean curvature, second fundamental form, etc) and the embedded curve (e.g. geodesic & normal curvatures, etc ). |
| + | |||
| + | The implementation uses a version of Sparse Field Level Sets (Whitaker et al 1998) modified for a mesh where there is an arbitrary number of vertex neighbors. | ||
| + | |||
| + | While the motivating problem is segmentation of Sulci & Gyri on the cortical surface, the technique is also applicable to analysis of other anatomic structures (e.g. bones and fractures thereof). | ||
== Usage == | == Usage == | ||
| − | === | + | ===Use Cases, Examples=== |
* Note use cases for which this module is especially appropriate, and/or link to examples. | * Note use cases for which this module is especially appropriate, and/or link to examples. | ||
* Link to examples of the module's use | * Link to examples of the module's use | ||
| − | * | + | |
| + | ===Tutorials=== | ||
| + | |||
| + | * Tutorial 1 | ||
| + | * Data Set 1 | ||
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
| − | |||
| + | A list panels in the interface, their features, what they mean, and how to use them. For instance: | ||
| + | |||
| + | {| | ||
| + | | | ||
* '''Input panel:''' | * '''Input panel:''' | ||
| + | ** '''First input''' | ||
| + | ** '''Second input''' | ||
* '''Parameters panel:''' | * '''Parameters panel:''' | ||
| + | ** '''First parameter''' | ||
| + | ** '''Second parameter''' | ||
* '''Output panel:''' | * '''Output panel:''' | ||
| + | ** '''First output''' | ||
| + | ** '''Second output''' | ||
* '''Viewing panel:''' | * '''Viewing panel:''' | ||
| + | |[[Image:screenshotBlankNotOptional.png|thumb|280px|User Interface]] | ||
| + | |} | ||
== Development == | == Development == | ||
| + | |||
| + | ===Notes from the Developer(s)=== | ||
| + | |||
| + | Algorithms used, library classes depended upon, use cases, etc. | ||
===Dependencies=== | ===Dependencies=== | ||
Other modules or packages that are required for this module's use. | Other modules or packages that are required for this module's use. | ||
| + | |||
| + | ===Tests=== | ||
| + | |||
| + | On the [http://www.cdash.org/CDash/index.php?project=Slicer3 Dashboard], these tests verify that the module is working on various platforms: | ||
| + | |||
| + | * MyModuleTest1 [http://viewvc.slicer.org/viewcvs.cgi/trunk MyModuleTest1.cxx] | ||
| + | * MyModuleTest2 [http://viewvc.slicer.org/viewcvs.cgi/trunk MyModuleTest2.cxx] | ||
===Known bugs=== | ===Known bugs=== | ||
| − | + | Links to known bugs in the Slicer3 bug tracker | |
| + | * [http://www.na-mic.org/Bug/view.php?id=000 Bug 000: description] | ||
| Line 61: | Line 92: | ||
===Source code & documentation=== | ===Source code & documentation=== | ||
| − | + | Links to the module's source code: | |
| − | |||
| − | |||
| + | Source code: | ||
| + | *[http://viewvc.slicer.org/viewcvs.cgi/trunk file.cxx ] | ||
| + | *[http://viewvc.slicer.org/viewcvs.cgi/trunk file.h ] | ||
| + | |||
| + | Doxygen documentation: | ||
| + | *[http://www.na-mic.org/Slicer/Documentation/Slicer3-doc/html/classes.html class1] | ||
== More Information == | == More Information == | ||
===Acknowledgment=== | ===Acknowledgment=== | ||
| − | + | This work was supported in part by grants from NSF, AFOSR, ARO, as well as by a grant from | |
| + | NIH (NAC P41 RR-13218) through Brigham and Women’s | ||
| + | Hospital. An NSF Fellowship supported part of the work. | ||
===References=== | ===References=== | ||
| − | + | ||
| + | P.A. Karasev, J.G. Malcolm, M. Niethammer, R. Kikinis, A. Tannenbaum. User-Driven 3D Mesh Region Targeting. To appear in SPIE Medical Imaging 2010. | ||
| + | [[File:pkmesh-spie.pdf]] | ||
Latest revision as of 20:19, 10 June 2010
Home < Modules:GyriContourSegmentation-Documentation-3.5Return to Slicer 3.5 Documentation
Mesh Contour Segmentation
MyModule
General Information
Module Type & Category
Type: CLI
Category: Segmentation
Authors, Collaborators & Contact
- Peter Karasev (Author): Georgia Tech
- Allen Tannenbaum (Collaborator): Georgia Tech
- Ron Kikinis (Collaborator): Harvard Medical School SPL
- Jimi Macolm (Collaborator): Harvard Medical School SPL / Georgia Tech
- Contact: Peter Karasev, pkarasev@gatech.edu
Module Description
This module is a tool to generate closed contours on a surface in 3D. The contour is initialized with a set of points, and subsequently 'evolves' according to some geometric criterion of the underlying surface (e.g. Surface Normal, mean curvature, second fundamental form, etc) and the embedded curve (e.g. geodesic & normal curvatures, etc ).
The implementation uses a version of Sparse Field Level Sets (Whitaker et al 1998) modified for a mesh where there is an arbitrary number of vertex neighbors.
While the motivating problem is segmentation of Sulci & Gyri on the cortical surface, the technique is also applicable to analysis of other anatomic structures (e.g. bones and fractures thereof).
Usage
Use Cases, Examples
- Note use cases for which this module is especially appropriate, and/or link to examples.
- Link to examples of the module's use
Tutorials
- Tutorial 1
- Data Set 1
Quick Tour of Features and Use
A list panels in the interface, their features, what they mean, and how to use them. For instance:
|
Development
Notes from the Developer(s)
Algorithms used, library classes depended upon, use cases, etc.
Dependencies
Other modules or packages that are required for this module's use.
Tests
On the Dashboard, these tests verify that the module is working on various platforms:
- MyModuleTest1 MyModuleTest1.cxx
- MyModuleTest2 MyModuleTest2.cxx
Known bugs
Links to known bugs in the Slicer3 bug tracker
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Links to the module's source code:
Source code:
Doxygen documentation:
More Information
Acknowledgment
This work was supported in part by grants from NSF, AFOSR, ARO, as well as by a grant from NIH (NAC P41 RR-13218) through Brigham and Women’s Hospital. An NSF Fellowship supported part of the work.
References
P.A. Karasev, J.G. Malcolm, M. Niethammer, R. Kikinis, A. Tannenbaum. User-Driven 3D Mesh Region Targeting. To appear in SPIE Medical Imaging 2010.