Difference between revisions of "Slicer3.4:Training"
From Slicer Wiki
| Line 72: | Line 72: | ||
= Slicer Tutorial Contest = | = Slicer Tutorial Contest = | ||
| − | |||
The following tutorials were part of the [http://www.na-mic.org/Wiki/index.php/Events:TutorialContestJune2009 June 2009 Slicer tutorial contest] and [http://www.na-mic.org/Wiki/index.php/Events:TutorialContestJan2009 January 2009 Slicer tutorial contest]. | The following tutorials were part of the [http://www.na-mic.org/Wiki/index.php/Events:TutorialContestJune2009 June 2009 Slicer tutorial contest] and [http://www.na-mic.org/Wiki/index.php/Events:TutorialContestJan2009 January 2009 Slicer tutorial contest]. | ||
| + | == '''June 2009 Tutorial Contest''' == | ||
{| border="1" cellpadding="5" | {| border="1" cellpadding="5" | ||
| Line 85: | Line 85: | ||
|- | |- | ||
| style="background:#849f66; color:#3b3f33; font-size:110%" align="Center"| <span id="1.1"></span> '''Confocal Microscopy''' | | style="background:#849f66; color:#3b3f33; font-size:110%" align="Center"| <span id="1.1"></span> '''Confocal Microscopy''' | ||
| − | | style="background:#CCFF99; color:black" | '''[http://www.na-mic.org/Wiki/images/4/4d/Microscopy-Confocal-TrainingTutorial-2009JUNE.pdf Confocal Microscopy] ''' | + | | style="background:#CCFF99; color:black" | '''[http://www.na-mic.org/Wiki/images/4/4d/Microscopy-Confocal-TrainingTutorial-2009JUNE.pdf Confocal Microscopy ] ''' |
| style="background:#CCFF99; color:black" | [http://www.ccdb.ucsd.edu/index.shtm Microscopy Confocal Dataset] | | style="background:#CCFF99; color:black" | [http://www.ccdb.ucsd.edu/index.shtm Microscopy Confocal Dataset] | ||
| style="background:#CCFF99; color:black" align="Center"| [[Image:MicroscopyTutorial.png| 250px]] | | style="background:#CCFF99; color:black" align="Center"| [[Image:MicroscopyTutorial.png| 250px]] | ||
| Line 108: | Line 108: | ||
| style="background:#CCFF99; color:black" | [http://www.na-mic.org/Wiki/index.php/File:LesionSegmentationTutorialData.zip Lesion Segmentation Tutorial Data] | | style="background:#CCFF99; color:black" | [http://www.na-mic.org/Wiki/index.php/File:LesionSegmentationTutorialData.zip Lesion Segmentation Tutorial Data] | ||
| style="background:#CCFF99; color:black" align="Center"| [[Image:LupusTutorial2.PNG| 250px]] | | style="background:#CCFF99; color:black" align="Center"| [[Image:LupusTutorial2.PNG| 250px]] | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | == '''January 2009 Slicer tutorial contest''' == | ||
| + | |||
| + | |||
| + | {| border="1" cellpadding="5" | ||
| + | |- style="background:#849f66; color:#3b3f33; font-size:130%" align="center" | ||
| + | | style="width:8%" ;align="Center"| '''Driving Biological Project/Collaboration''' | ||
| + | | style="width:40%" ;align="Center"| '''Tutorial''' | ||
| + | | style="width:40%" ;align="Center"| '''Sample Data''' | ||
| + | | style="width:12%" ;align="Center"| '''Image''' | ||
|- | |- | ||
| style="background:#849f66; color:#3b3f33; font-size:110%" align="Center"| <span id="1.1"></span> '''Automated FE Mesh''' | | style="background:#849f66; color:#3b3f33; font-size:110%" align="Center"| <span id="1.1"></span> '''Automated FE Mesh''' | ||
Revision as of 14:44, 5 May 2010
Home < Slicer3.4:TrainingContents
Slicer 3.4 Tutorials
- The following table contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version 3.4) to accomplish certain tasks.
- For questions related to the Slicer3.4 Compendium, please send an e-mail to Sonia Pujol, Ph.D. (spujol at bwh.harvard.edu).
| Category | Tutorial | Sample Data | Image |
| Basic | Slicer3Minute Tutorial The Slicer3Minute tutorial is an introduction to the advanced 3D visualization capabilities of Slicer3.4. Audience: First time users. |
Slicer3Minute dataset The Slicer3Minute dataset contains an MR scan of the brain and 3D reconstructions of the anatomy |
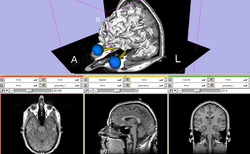
|
| Basic | Slicer3 Manual Registration Tutorial shows how to manually/interactively align two images in Slicer3.4 or 3.5 Audience: First time & early users. |
Manual Registration example dataset This dataset contains two brain MRI of a single subject, obtained in different orientations. |
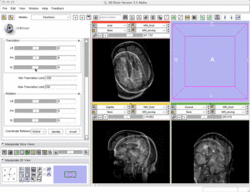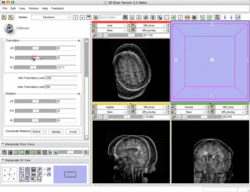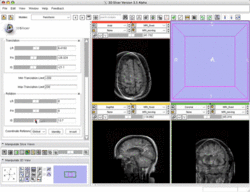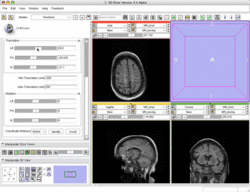
|
| Core | Slicer3Visualization Tutorial The Slicer3Visualization tutorial guides through 3D data loading and visualization in Slicer3.4. Audience: All beginning users including clinicians, scientists, engineers and programmers. |
Slicer3Visualization dataset The Slicer3VisualizationDataset contains two MR scans of the brain, a pre-computed labelmap and 3D reconstructions of the anatomy. |
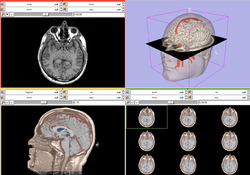
|
| Core | Programming in Slicer3 Tutorial The Programming in Slicer3 tutorial is an introduction to the the integration of stand-alone programs outside of the Slicer3 source tree. Audience: Programmers and Engineers. |
HelloWorld Plugin The HelloWorld tutorial dataset contains an MR scan of the brain and pre-computed xml and C++ files for integrating the Hello World plug-in to Slicer3. |
|
| Specialized | 3D Visualization of FreeSurfer Data The course guides through 3D visualization of FreeSurfer brain segmentations, surface reconstruction and parcellation results in Slicer3.4. Audience: All users. |
FreeSurfer Tutorial dataset The FreeSurfer dataset contains an MR scan of the brain and pre-computed FreeSurfer segmentation and cortical surface reconstructions. |
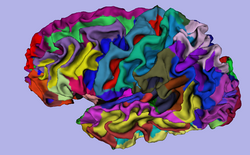
|
| Specialized | Automatic Segmentation Tutorial The course guides through the process of using the Expectation-Maximization Segmentation algorithm to automatically segment brain structures from MRI data. Audience: Programmers and Engineers. |
Automatic Segmentation dataset | 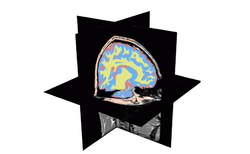
|
| Specialized | Atlas Label Merging & Surface Based Registration Tutorial This tutorial guides through the creation and co-registration of surface models of atlas structures (thalamus & thalamic nuclei) and subsequent merging of two labelmaps. |
Atlas Merging tutorial dataset (contains 2 full brain atlases + intermediate result files, 72MB) | 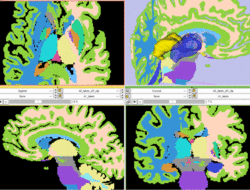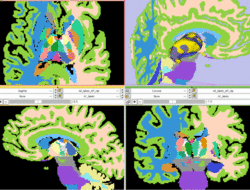
|
| Specialized | Neurosurgical Planning Tutorial This tutorial takes the trainee through a complete workup for neurosurgical patient-specific mapping. Also see this tutorial for information on how to use Slicer's affine registration, simple region growing, model maker and tractography modules. Audience: All users interested in image-guided therapy. |
Neurosurgical Planning dataset | 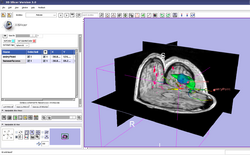
|
| Specialized | Diffusion MRI Tutorial This tutorial guides you through the process of loading diffusion MR data, estimating diffusion tensors, and performing tractography of white matter bundles. Audience: All users and developers. |
Diffusion dataset | 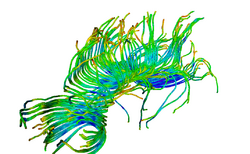
|
| Specialized | ChangeTracker Tutorial This tutorial describes the use of ChangeTracker module to detect changes in tumor volume from two MRI scans. Audience: All users interested in longitudinal analysis of pathology. |
Training data download is integrated with the ChangeTracker module (see Tutorial) | |
| Specialized | LiverSegmentation Tutorial This tutorial introduces translational clinical scientists to the capabilities of the 3D Slicer software through the segmentation of the liver. Audience: All users and developers. |
Liver Segmentation dataset | 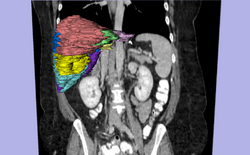
|
Slicer Tutorial Contest
The following tutorials were part of the June 2009 Slicer tutorial contest and January 2009 Slicer tutorial contest.
June 2009 Tutorial Contest
| Driving Biological Project/Collaboration | Tutorial | Sample Data | Image |
| Confocal Microscopy | Confocal Microscopy | Microscopy Confocal Dataset | 
|
| Autism | ARCTIC: Automatic Regional Cortical ThICkness V2.1 | ARCTIC Data | 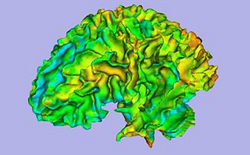
|
| Prostate | Trans-rectal MR guided prostate biopsy | MR guided prostate biopsy Dataset | 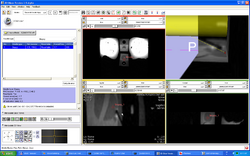
|
| Schizophrenia | Python Stochastic Tractography Module | Stochastic Tractography Data | 
|
| Lupus | White Matter Lesions Segmentation V2.2 | Lesion Segmentation Tutorial Data | 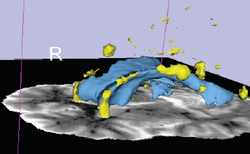
|
January 2009 Slicer tutorial contest
| Driving Biological Project/Collaboration | Tutorial | Sample Data | Image |
| Automated FE Mesh | IA FEMesh Tutorial | Mesh Tutorial Example Data | 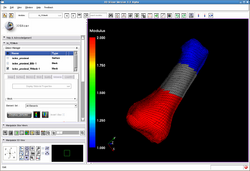
|
| Alcohol and Stress Interaction | Non-Human Primate Segmentation Tutorial | Non-Human Primate Segmentation Data | 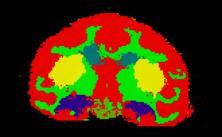
|
| Prostate | MR Guided Prostate Interventions | MR Guided Prostate Interventions Data | 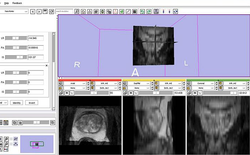
|
| IGT | Basic Navigation Tutorial | Basic Navigation Data | 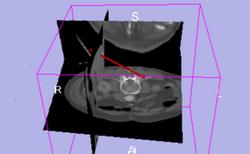
|
| Schizophrenia | Python Stochastic Tractography Tutorial V1.0 | Stochastic Tractography Data | 
|
| Autism | ARCTIC: Automatic Regional Cortical ThICkness V1.0 | ARCTIC Data | 
|
| Lupus | White Matter Lesions Segmentation V2.0 | Lesion Segmentation Tutorial Data | 
|
| Orbit Segmentation | Segmentation of the Orbit | Orbit Segmentation Data | 
|
| VMTK | VMTK Coronary Arteries Centrelines Extraction | VMTK Centrelines Data | 
|
| Fiber Clustering | EM Fiber Clustering | EM Fiber Clustering Data | 
|
Slicer 3.5 Tutorials
- The following table contains "How to" tutorials with matched sample data sets. They demonstrate how to use the 3D Slicer environment (version 3.5) to accomplish certain tasks.
| Contest | Tutorial | Sample Data | Image |
| Specialized | Robust Statistics Segmentation Tutorial | Robust Statistics Segmentation dataset | 
|
Software Installation
- The Slicer download page contains information on how to obtain a compiled version of Slicer for a variety of platforms and where to find the source code for Slicer 3.
Software Documentation
- For the Slicer 3.4 manual pages please click here. These pages are the reference manual for Slicer 3.4 and briefly explain the functionality found in panels and modules.
Older Tutorials
- Visit the Slicer 3.2 training page.
- Visit the Slicer 2 training page.