Difference between revisions of "Modules:NeuroNav-Documentation-3.6"
| (27 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Documentation-3.6|Return to Slicer 3.6 Documentation]] | [[Documentation-3.6|Return to Slicer 3.6 Documentation]] | ||
| − | [[Announcements:Slicer3. | + | [[Announcements:Slicer3.4#Highlights|Gallery of New Features]] |
__NOTOC__ | __NOTOC__ | ||
| Line 24: | Line 24: | ||
* Haiying Liu, BWH | * Haiying Liu, BWH | ||
* Nobuhiko Hata, BWH | * Nobuhiko Hata, BWH | ||
| + | * Ron Kikinis, BWH | ||
* Contact: Haiying Liu, hliu@bwh.harvard.edu | * Contact: Haiying Liu, hliu@bwh.harvard.edu | ||
===Module Description=== | ===Module Description=== | ||
| − | NeuroNav module is developed to guide neurosurgeons by images to perform surgical procedures in OR | + | NeuroNav module is developed to guide neurosurgeons by images to perform surgical procedures in OR. |
| − | |||
| − | |||
| − | |||
== Usage == | == Usage == | ||
| + | ===Regular Neurosurgical Navigation=== | ||
| + | Follow the following steps to make the module work properly: | ||
| + | #Connect to a tracking source, which could be a tracking simulator or real tracking device such as NDI Aurora. The tracking source sends tracking data, i.e. the orientation and tip location of the probe, to Slicer at a defined rate. | ||
| + | #Perform patient to image registration. Load preoperative image(s) or scene into Slicer. Choose 4-6 landmarks on the patient head and then find those points on the loaded image to perform the patient to image registration. | ||
| + | #Execute surgical navigation. Navigation is driven by the tracking probe. As long as the probe tip moves within the tracking scope of the device, Slicer will update the image display to give a visual feedback to the neurorsurgeon. The image update will offer useful real-time guidance during the surgical procedure. | ||
| − | === | + | ===Integration with BioImage Suite and BrainLab=== |
| + | Integration of Slicer with BioImage Suite and BrainLab has been a recent successful example of collaboration between academia and industry. | ||
| + | BioImage Suite is an integrated image analysis software suite developed at Yale University. BrainLab is an FDA approved neurosurgical navigation system. This integration will interface Slicer to the BrainLab system and allow neurosurgeons to use Slicer to perform some research in DTI visualization in OR while using BrainLab as the navigation system. Please check sections below for details. | ||
| − | + | ===Examples, Use Cases & Tutorials=== | |
| − | + | [[Slicer3:BrainLab_Integration|Tutorial for the integration of Slicer with BioImage Suite and BrainLab system.]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | ===Tutorials=== | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
| − | + | * '''Connection:''' Connect to a tracking source through OpenIGTLink. This step is not visible on NeuroNav module interface. | |
| − | + | * '''Registration:''' Perform patient to image registration. | |
| − | + | * '''Navigation:''' Execute surgical navigation. | |
| − | |||
| − | * ''' | ||
| − | |||
| − | |||
| − | * ''' | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | * ''' | ||
| − | |||
| − | |||
== Development == | == Development == | ||
===Notes from the Developer(s)=== | ===Notes from the Developer(s)=== | ||
| − | |||
| − | |||
===Dependencies=== | ===Dependencies=== | ||
| − | + | Fiducial<br> | |
| − | + | OpenIGTLink<br> | |
| + | DTMRI<br> | ||
| + | FiducialSeeding<br> | ||
===Tests=== | ===Tests=== | ||
| − | On the [http://www.cdash.org/CDash/index.php?project=Slicer3 Dashboard], these tests verify that the module is working on various platforms | + | On the [http://www.cdash.org/CDash/index.php?project=Slicer3 Dashboard], these tests verify that the module is working on various platforms. |
| − | |||
| − | |||
| − | |||
===Known bugs=== | ===Known bugs=== | ||
| − | + | Follow this [http://na-mic.org/Mantis/main_page.php link] to the Slicer3 bug tracker. | |
| − | |||
| − | |||
| − | |||
| − | |||
===Usability issues=== | ===Usability issues=== | ||
| Line 101: | Line 72: | ||
===Source code & documentation=== | ===Source code & documentation=== | ||
| + | Follow this [http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/Volumes/ link] to the Volumes source code in ViewVC. | ||
| − | + | [http://www.na-mic.org/Slicer/Documentation/Slicer3-doc/html/classes.html Documentation] generated by doxygen. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== More Information == | == More Information == | ||
===Acknowledgment=== | ===Acknowledgment=== | ||
| − | + | This work is part of the National Center for Image Guided Therapy (NCIGT), funded by the National Institutes of Health, Grant 5U41RR019703. This study is also in part supported by Intelligent Surgical Instruments Project of METI (Japan). | |
===References=== | ===References=== | ||
| − | + | H Elhawary, Haiying Liu, Pratik Patel, Isaiah Norton, Laura Rigolo, Xenophon Papademetris, Nobuhiko Hata, Alexandra J. Golby, Intra-operative Real-time Querying of White Mater Tracts during Frameless Stereotactic Neuronavigation, in press, Neurosurgery, 2010 | |
Latest revision as of 13:55, 29 April 2010
Home < Modules:NeuroNav-Documentation-3.6Return to Slicer 3.6 Documentation
Module Name
NeuroNav
General Information
Module Type & Category
Type: Interactive
Category: IGT
Authors, Collaborators & Contact
- Haiying Liu, BWH
- Nobuhiko Hata, BWH
- Ron Kikinis, BWH
- Contact: Haiying Liu, hliu@bwh.harvard.edu
Module Description
NeuroNav module is developed to guide neurosurgeons by images to perform surgical procedures in OR.
Usage
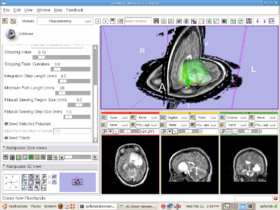
Follow the following steps to make the module work properly:
- Connect to a tracking source, which could be a tracking simulator or real tracking device such as NDI Aurora. The tracking source sends tracking data, i.e. the orientation and tip location of the probe, to Slicer at a defined rate.
- Perform patient to image registration. Load preoperative image(s) or scene into Slicer. Choose 4-6 landmarks on the patient head and then find those points on the loaded image to perform the patient to image registration.
- Execute surgical navigation. Navigation is driven by the tracking probe. As long as the probe tip moves within the tracking scope of the device, Slicer will update the image display to give a visual feedback to the neurorsurgeon. The image update will offer useful real-time guidance during the surgical procedure.
Integration with BioImage Suite and BrainLab
Integration of Slicer with BioImage Suite and BrainLab has been a recent successful example of collaboration between academia and industry. BioImage Suite is an integrated image analysis software suite developed at Yale University. BrainLab is an FDA approved neurosurgical navigation system. This integration will interface Slicer to the BrainLab system and allow neurosurgeons to use Slicer to perform some research in DTI visualization in OR while using BrainLab as the navigation system. Please check sections below for details.
Examples, Use Cases & Tutorials
Tutorial for the integration of Slicer with BioImage Suite and BrainLab system.
Quick Tour of Features and Use
- Connection: Connect to a tracking source through OpenIGTLink. This step is not visible on NeuroNav module interface.
- Registration: Perform patient to image registration.
- Navigation: Execute surgical navigation.
Development
Notes from the Developer(s)
Dependencies
Fiducial
OpenIGTLink
DTMRI
FiducialSeeding
Tests
On the Dashboard, these tests verify that the module is working on various platforms.
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Follow this link to the Volumes source code in ViewVC.
Documentation generated by doxygen.
More Information
Acknowledgment
This work is part of the National Center for Image Guided Therapy (NCIGT), funded by the National Institutes of Health, Grant 5U41RR019703. This study is also in part supported by Intelligent Surgical Instruments Project of METI (Japan).
References
H Elhawary, Haiying Liu, Pratik Patel, Isaiah Norton, Laura Rigolo, Xenophon Papademetris, Nobuhiko Hata, Alexandra J. Golby, Intra-operative Real-time Querying of White Mater Tracts during Frameless Stereotactic Neuronavigation, in press, Neurosurgery, 2010