Difference between revisions of "Modules:RobustStatisticsSeg-Documentation-3.6"
(Created page with 'Return to Slicer 3.6 Documentation __NOTOC__ ===Robust Statistics Segmentation=== {| |thumb|280px|Segmentation result |} ==…') |
|||
| Line 4: | Line 4: | ||
{| | {| | ||
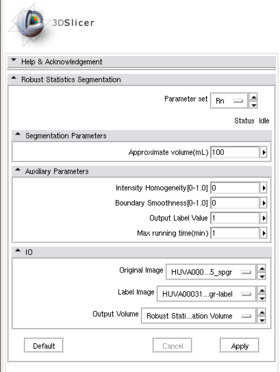
| − | |[[Image:RSS_MultiObjSeg1.png|thumb| | + | |[[Image:RssPanel.png|thumb|280px|RSS module panel]] |
| + | |[[Image:RSS_MultiObjSeg1.png|thumb|300px|RSS result]] | ||
|} | |} | ||
Revision as of 23:06, 7 April 2010
Home < Modules:RobustStatisticsSeg-Documentation-3.6Return to Slicer 3.6 Documentation
Robust Statistics Segmentation
General Information
Module Type & Category
Type: CLI
Category: Segmentation
Authors, Collaborators & Contact
- Yi Gao (Author): Georgia Tech
- Allen Tannenbaum (Collaborator): Georgia Tech
- Ron Kikinis (Collaborator): BWH
- Contact: Yi Gao, yi.gao@gatech.edu
Module Description
This module is a general purpose segmentor. The target object is initialized by a label map. An active contour model then evolves to extract the desired boundary of the object.
Usage
Use Cases, Examples
- Note use cases for which this module is especially appropriate, and/or link to examples.
- Link to examples of the module's use
Tutorials
- Tutorial 1
- Data Set 1
Quick Tour of Features and Use
A list panels in the interface, their features, what they mean, and how to use them. For instance:
|
Development
Notes from the Developer(s)
Algorithms used, library classes depended upon, use cases, etc.
Dependencies
Other modules or packages that are required for this module's use.
Tests
On the Dashboard, these tests verify that the module is working on various platforms:
- MyModuleTest1 MyModuleTest1.cxx
- MyModuleTest2 MyModuleTest2.cxx
Known bugs
Links to known bugs in the Slicer3 bug tracker
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Links to the module's source code:
Source code:
Doxygen documentation:
More Information
Acknowledgment
This work was supported in part by grants from NSF, AFOSR, ARO, as well as by a grant from NIH (NAC P41 RR-13218) through Brigham and Women’s Hospital. An NSF Fellowship supported part of the work.